Abstract
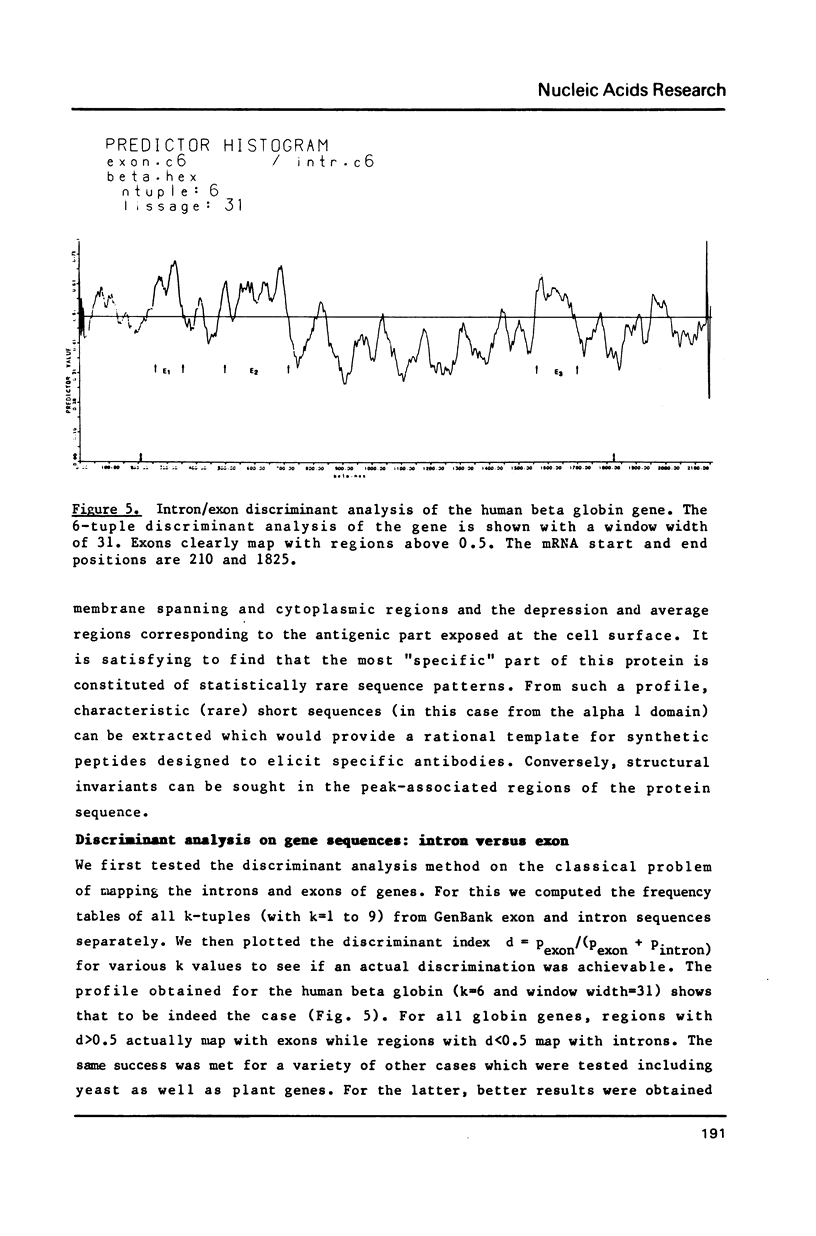
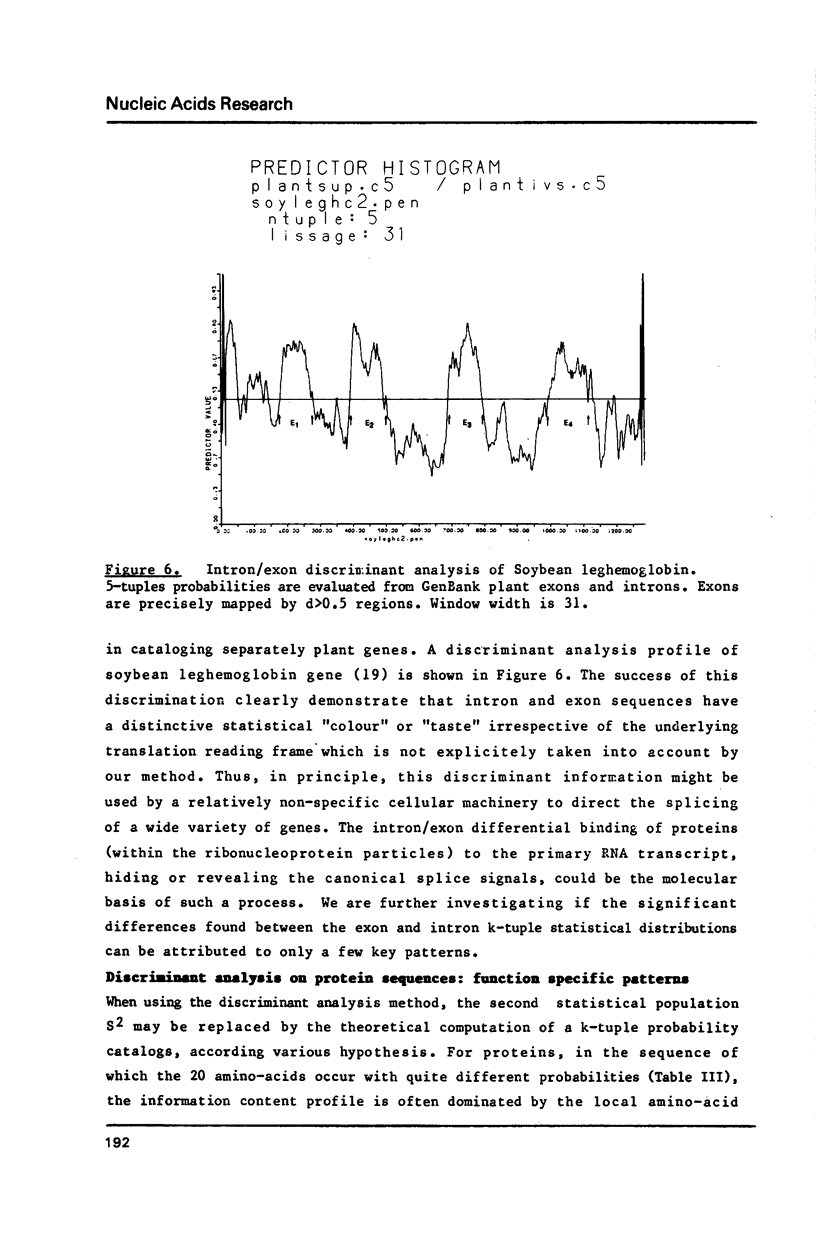
Nucleotide or amino-acid sequences are interpreted as successions of words of length k (k-tuples) the frequencies of which are highly variable in different statistical populations of genes or proteins. After building k-tuple reference tables from coherent subsets or entire data banks, the local information content profile of individual sequences is drawn. Anomalous regions (peaks or depressions) of such a profile can lead to the discovery and identification of specific sequence patterns. Along the same principle, the simultaneous use of two reference statistical populations and the computation of an index combining the two information profiles lead to a general and powerful discriminant analysis methods. The identification of a "signal" associated with gene conversion, the introns/exons discrimination and the location of function specific patterns in proteins are given as examples of successful applications of this heuristic informational approach.
Full text
PDF











Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arnold B., Burgert H. G., Archibald A. L., Kvist S. Complete nucleotide sequence of the murine H-2Kk gene. Comparison of three H-2K locus alleles. Nucleic Acids Res. 1984 Dec 21;12(24):9473–9487. doi: 10.1093/nar/12.24.9473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bach R., Iwasaki Y., Friedland P. Intelligent computational assistance for experiment design. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):11–29. doi: 10.1093/nar/12.1part1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A. P. DNA methylation and the frequency of CpG in animal DNA. Nucleic Acids Res. 1980 Apr 11;8(7):1499–1504. doi: 10.1093/nar/8.7.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop M., Thompson E. Fast computer search for similar DNA sequences. Nucleic Acids Res. 1984 Jul 11;12(13):5471–5474. doi: 10.1093/nar/12.13.5471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brisson N., Verma D. P. Soybean leghemoglobin gene family: normal, pseudo, and truncated genes. Proc Natl Acad Sci U S A. 1982 Jul;79(13):4055–4059. doi: 10.1073/pnas.79.13.4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucher P., Bryan B. Signal search analysis: a new method to localize and characterize functionally important DNA sequences. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):287–305. doi: 10.1093/nar/12.1part1.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claverie J. M. A common philosophy and FORTRAN 77 software package for implementing and searching sequence databases. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):397–407. doi: 10.1093/nar/12.1part1.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claverie J. M., Sauvaget I. Assessing the biological significance of primary structure consensus patterns using sequence databanks. I. Heat-shock and glucocorticoid control elements in eukaryotic promoters. Comput Appl Biosci. 1985;1(2):95–104. doi: 10.1093/bioinformatics/1.2.95. [DOI] [PubMed] [Google Scholar]
- Claverie J. M., Sauvaget I., Bougueleret L. Computer generation and statistical analysis of a data bank of protein sequences translated from GenBank. Biochimie. 1985 May;67(5):437–443. doi: 10.1016/s0300-9084(85)80261-3. [DOI] [PubMed] [Google Scholar]
- Dumas J. P., Ninio J. Efficient algorithms for folding and comparing nucleic acid sequences. Nucleic Acids Res. 1982 Jan 11;10(1):197–206. doi: 10.1093/nar/10.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gribskov M., Devereux J., Burgess R. R. The codon preference plot: graphic analysis of protein coding sequences and prediction of gene expression. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 2):539–549. doi: 10.1093/nar/12.1part2.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlin S., Ghandour G., Ost F., Tavare S., Korn L. J. New approaches for computer analysis of nucleic acid sequences. Proc Natl Acad Sci U S A. 1983 Sep;80(18):5660–5664. doi: 10.1073/pnas.80.18.5660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korn L. J., Queen C. Analysis of biological sequences on small computers. DNA. 1984 Dec;3(6):421–436. doi: 10.1089/dna.1.1984.3.421. [DOI] [PubMed] [Google Scholar]
- Kourilsky P. Genetic exchanges between partially homologous nucleotide sequences: possible implications for multigene families. Biochimie. 1983 Feb;65(2):85–93. doi: 10.1016/s0300-9084(83)80178-3. [DOI] [PubMed] [Google Scholar]
- Lipman D. J., Pearson W. R. Rapid and sensitive protein similarity searches. Science. 1985 Mar 22;227(4693):1435–1441. doi: 10.1126/science.2983426. [DOI] [PubMed] [Google Scholar]
- Lyall A., Hammond P., Brough D., Glover D. BIOLOG - a DNA sequence analysis system in PROLOG. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 2):633–642. doi: 10.1093/nar/12.1part2.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orcutt B. C., George D. G., Dayhoff M. O. Protein and Nucleic Acid Sequence Database Systems. Annu Rev Biophys Bioeng. 1983;12:419–441. doi: 10.1146/annurev.bb.12.060183.002223. [DOI] [PubMed] [Google Scholar]
- Staden R. Graphic methods to determine the function of nucleic acid sequences. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 2):521–538. doi: 10.1093/nar/12.1part2.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilbur W. J., Lipman D. J. Rapid similarity searches of nucleic acid and protein data banks. Proc Natl Acad Sci U S A. 1983 Feb;80(3):726–730. doi: 10.1073/pnas.80.3.726. [DOI] [PMC free article] [PubMed] [Google Scholar]



