Rules of the genetic code can be temporarily suspended to make room for a variety of specific tricks that direct synthesis of additional proteins from mRNAs that would not be predicted by their sequences. This “recoding” includes specification of the 21st encoded amino acid, selenocysteine (1, 2). In another form, recoding itself provides a sensing mechanism to regulate expression (3). In other cases mRNA can direct synthesis of a set ratio of two proteins that share some amino acid sequences. Kim, Su, Maas, O’Neill, and Rich, in this issue of PNAS (4), describe experiments that show the mRNA structural features important for one such case of ratio setting.
We know little about the global contribution that recoding makes to the complexity of proteomes. It could be that many mRNAs encode, in addition to their standard products, minor products that so far would have escaped detection. These products are likely to have distinct functions and contribute to the biological complexity of the proteome. Clearly the complexity of the human proteome is far beyond the more than 100,000 human genes. The total mRNA population might be 250,000 from alternate splicing, editing, and use of alternate promoters. How much this complexity will be expanded by protein modification, protein splicing, and recoding is far from clear but a total of 500,000 might not be surprising. Certainly the contribution from recoding is completely unknown; however, specific examples being studied give some guide to what is to come.
Recoding by redefinition of codons and translational bypassing are being studied in detail but the majority of known examples are programmed shifts in reading frame (3, 5). Depending on the shift site ribosomes can be instructed to slip into +1 or the −1 frame. Examples are found in all well-studied organisms from viruses to bacteria to humans, but in the case of human gene expression only one family of genes, antizyme, is known to use frameshifting and this is +1 (6, 7). Many more cases of −1 frameshifting are known partly because of their use by members of large families of viruses and bacterial insertion sequences. Programmed −1 frameshifting occurs at “slippery” shift sites. At these sites, initial codon-anticodon pairing in the zero frame is followed by dissociation and re-pairing of the tRNA anticodon to mRNA at an overlapping codon. The great majority of cases involve realignment of the codon anticodon pairing of not just one tRNA but of two adjacent tRNAs in the ribosome (peptidyl tRNA and aminoacyl tRNA). Even though the pairing rules for re-pairing are more relaxed than those for initial pairing, tandem slippage gives signature shift sites of the general form X XXY YYZ where X, Y, and Z can even be the same nucleotides. Tandem slippage was first recognized for the programmed frameshifting that occurs close to the 3′ end of the gag gene of many retroviruses (8). This frameshifting is required for synthesis of the GagPol fusion polyprotein as there is no independent ribosome entry to the pol gene. [In retroviruses that have a separate protease gene between gag and pol two frameshift events are required to synthesize the GagProPol fusion polyprotein (9, 10).]
The potential for codon:anticodon alignment is not the only important feature for programmed −1 frameshifting. Frameshifting is stimulated by discrete mRNA structures that in nearly all cases are formed from contiguous sequence starting 5–9 nt 3′ of the shift site. Although in HIV (11) and Escherichia coli dnaX (12, 13) this stimulating structure is a simple stem loop, and in IS911 it is a three-way junction (14), in most cases it is a pseudoknot as first recognized in studies on viruses (15–17).
There are various types of pseudoknots, but the classical H-(hairpin) pseudoknot (18) requires pairing of the loop of a hairpin loop with downstream sequences, resulting in two stems that are connected by two loops. RNA pseudoknots are widely occurring structural motifs that are involved in various RNA functions (19). Pseudoknots were first recognized experimentally from studying the folding of the 3′ end of the turnip yellow mosaic virus RNA (for review see ref. 20). They have since been found in many classes of RNA including: ribosomal RNA (21), catalytic and self-splicing RNA (22, 23), tmRNA (24–26), internal ribosome entry sites of some picornaviruses (27), and translational repression sites on mRNAs for some ribosomal proteins (28) and for certain T-even bacteriophage proteins (29, 30). However, the pseudoknots involved in recoding are unique in that, as they play their role as a structure, they are immediately unfolded and their now linear sequence serves as a template for decoding.
Pseudoknots cause ribosomal pausing, raising the possibility that this delay at the crucial site allows more time for anticodon:mRNA realignment to occur. This hypothesis has been tested in two cases with the conclusion that pausing may be necessary for −1 frameshifting but it is not sufficient (31, 32)—something else must be going on. Much remains to be learned, including when the pseudoknot is melted out and with what the pseudoknot might interact (see ref. 33 for discussion).
Kim et al. (4) describe experiments that functionally test the unique structural features of a frameshift stimulating pseudoknot from the plant virus beet western yellow virus (BWYV), a luteovirus. Replication of BYMV genomic RNA requires two virus-encoded functions, P1 and P2 (for review see ref. 34). The polymerase, P2, is expressed only as an extension of P1 when a few percent of the ribosomes, three-quarters of the way through the coding sequence for P1, shift reading frame into the overlapping ORF for P2 (16, 35, 36). This means that the P1 protein and the P1-P2 frameshifted protein share their first 461 aa. Then, each uses a different frame to decode the next 146 aa, and P1-P2 then continues for another 429 unique aa. The frameshift event is a −1 tandem shift at a G GGG AAC site (the same shift site used by simian retrovirus 1; ref. 37), stimulated by a pseudoknot 6 nt downstream (Fig. 1).
Figure 1.
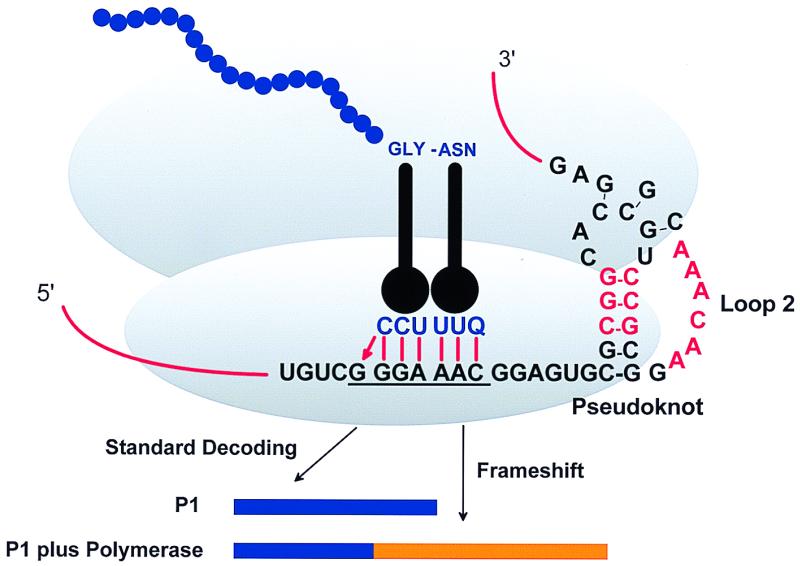
Model for BWYV −1 frameshift. Bases in red are conserved in all known luteoviruses.
Rich’s group (38) recently described a crystal structure of the BWYV pseudoknot. The structure shows that the two stems are not coaxially stacked because of a 48° rotation between the two pairs of bases at the junction and because of unusual conformations of the junctional U and A bases. The result is an overall bend angle between the stems of 25°. There is a quadruple base interaction between a nucleotide in loop 1 and stem regions. And perhaps most significantly loop 2 forms a triple helix through interactions with the minor groove of stem 1. The experiments reported by Kim et al. (4) test the functional importance of key nucleotides in this structure (38) for stimulation of frameshifting.
The mutational results support the complex structure—residues predicted to have key interactions are crucial for activity and those that are not interacting are insensitive to change. The unusual structural features of the interactions between loop 2 and stem 1 are functionally important as are the specific interactions around the stem junctions. A few bases are unexpectedly sensitive to mutation and the suggestion is that these may make key contacts with ribosomes or perhaps some other factor important to the process. Interestingly, some mutational changes increase the relatively low efficiency of this shift site to a higher level, implying that selection has led to a system tuned to the most useful efficiency.
From these results, and the other well-studied examples of pseudoknots that promote frameshifting, it is clear that more than one structure can suffice (39–41) but not all pseudoknots work (42).
The differences between active pseudoknots seem particularly apparent in the importance of loop 2 sequences. In BWYV, loop 2 plays a crucial structural and functional role. With infectious bronchitis virus (17) and simian retrovirus-1 (37, 43) pseudoknots, loop 2 sequences have great latitude as if their role is merely to connect stems. However, identities of some bases are crucial in loop 2 of the pseudoknot in murine leukemia virus that stimulates, not frameshifting, but in-frame read-through of a stop codon. (The mouse mammary tumor virus pseudoknot cannot substitute for its murine leukemia virus counterpart in stimulating read-through; ref. 44.) Presumably, in the cases where the identities of loop 2 bases are unimportant, the sequences are not forming part of an essential structure, nor are they forming key interactions with ribosomes or other possible factors.
Both the structure and the function studies with BWYV point to the crucial role of the stem-junction region. This is also the case with the mouse mammary tumor virus pseudoknot, but here the junction is very different. The NMR-based structure shows an angle of 60° between the stems. This is caused by a “wedge” base on one side at the junction (39, 45, 46), and functional tests imply that this base, and hence the bend, is important (47).
However, it would be comforting and perhaps even instructive to have both x-ray and NMR structures for the same pseudoknot.
The spacer region between the hepta-nucleotide shift site and the pseudoknot is also important for function. Changing the spacer length generally decreases frameshifting. Presumably the spacer length alters the position of the paused ribosome and influences the probability of shifting. Sequence identity of the spacer has not been sufficiently tested for frameshifting cases. It is known that the identity of some nucleotides is essential for pseudoknot-dependent read-through in murine leukemia virus (44, 48)—these bases could be interacting either with the pseudoknot to form a more complicated structure or perhaps with translational components. The spacer sequences in BWYV should be tested both for functional importance and for possible influences on the pseudoknot structure. At the same time it would be worth testing to make sure that the stop codon inserted for convenience immediately after the shift site does not influence the results (see ref. 49).
The crystal structure of the BWYV combined with the new mutational results clearly shows the complexity and sophistication of RNA structures that in this case form transiently to promote alteration of decoding. This is now a firm footing from which to determine just how this structural information of pseudoknots is used to influence the mechanism of frameshifting. It is equally a challenge to discover the exploitation of these mechanisms for expression of the mammalian proteome.
Footnotes
See companion article on page 14234.
References
- 1.Baron C, Böck A. In: tRNA, Structure, Biosynthesis, and Function. Söll D, RajBhandary U L, editors. Washington DC: Am. Soc. Microbiol.; 1995. pp. 529–544. [Google Scholar]
- 2.Low S C, Berry M J. Trends Biochem Sci. 1996;21:203–208. [PubMed] [Google Scholar]
- 3.Gesteland R F, Atkins J F. Annu Rev Biochem. 1996;65:741–768. doi: 10.1146/annurev.bi.65.070196.003521. [DOI] [PubMed] [Google Scholar]
- 4.Kim Y-G, Su L, Maas S, O’Neill A, Rich A. Proc Natl Acad Sci USA. 1999;96:14234–14239. doi: 10.1073/pnas.96.25.14234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Atkins J F, Böck A, Matsufuji S, Gesteland R F. In: The RNA World. 2nd Ed. Gesteland R F, Cech T R, Atkins J F, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1999. pp. 637–673. [Google Scholar]
- 6.Matsufuji S, Matsufuji T, Miyazaki Y, Murakami Y, Atkins J F, Gesteland R F, Hayashi S. Cell. 1995;80:51–60. doi: 10.1016/0092-8674(95)90450-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ivanov I P, Gesteland R F, Atkins J F. Genomics. 1998;52:119–129. doi: 10.1006/geno.1998.5434. [DOI] [PubMed] [Google Scholar]
- 8.Jacks T, Madhani H D, Masiarz R F, Varmus H E. Cell. 1988;55:447–458. doi: 10.1016/0092-8674(88)90031-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moore R, Dixon M, Smith R, Peters G, Dickson C. J Virol. 1987;61:480–490. doi: 10.1128/jvi.61.2.480-490.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jacks T, Townsley K, Varmus H E, Majors J. Proc Natl Acad Sci USA. 1987;84:4298–4302. doi: 10.1073/pnas.84.12.4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parkin N T, Chamorro M, Varmus H E. J Virol. 1992;66:5147–5151. doi: 10.1128/jvi.66.8.5147-5151.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsuchihashi Z. Nucleic Acids Res. 1991;19:2457–2462. doi: 10.1093/nar/19.9.2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Larsen B, Gesteland R F, Atkins J F. J Mol Biol. 1997;271:47–60. doi: 10.1006/jmbi.1997.1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rettberg C C, Prère M F, Gesteland R F, Atkins J F, Fayet O. J Mol Biol. 1999;286:1365–1378. doi: 10.1006/jmbi.1999.2546. [DOI] [PubMed] [Google Scholar]
- 15.Brierley I, Digard P, Inglis S C. Cell. 1989;57:537–547. doi: 10.1016/0092-8674(89)90124-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.ten Dam E B, Pleij C W A, Bosch L. Virus Genes. 1990;4:121–136. doi: 10.1007/BF00678404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brierley I, Rolley N J, Jenner A J, Inglis S C. J Mol Biol. 1991;220:889–902. doi: 10.1016/0022-2836(91)90361-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gultyaev A, van Batenburg F H D, Pleij C W A. RNA. 1999;5:609–617. doi: 10.1017/s135583829998189x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deiman B A L M, Pleij C W A. Semin Virol. 1997;8:166–175. [Google Scholar]
- 20.Pleij C W A, Rietveld K, Bosch L. Nucleic Acids Res. 1985;13:1717–1731. doi: 10.1093/nar/13.5.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moazed D, Noller H F. Nature (London) 1987;327:389–394. doi: 10.1038/327389a0. [DOI] [PubMed] [Google Scholar]
- 22.Cate J H, Gooding A R, Podell E, Zhou K, Golden B L, Kundrot C E, Cech T R, Doudna J A. Science. 1996;273:1678–1685. doi: 10.1126/science.273.5282.1678. [DOI] [PubMed] [Google Scholar]
- 23.Ferré-D’Amaré A R, Zhou K H, Doudna J A. Nature (London) 1998;395:567–574. doi: 10.1038/26912. [DOI] [PubMed] [Google Scholar]
- 24.Williams K P, Bartel D P. RNA. 1996;2:1306–1310. [PMC free article] [PubMed] [Google Scholar]
- 25.Felden B, Himeno H, Muto A, McCutcheon J P, Atkins J F, Gesteland R F. RNA. 1997;3:89–103. [PMC free article] [PubMed] [Google Scholar]
- 26.Nameki N, Chattopadhyay P, Himeno H, Muto A, Kawai G. Nucleic Acids Res. 1999;27:3667–3675. doi: 10.1093/nar/27.18.3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rijnbrand R, van der Straaten T, van Rijn P A, Spaan W J, Bredenbeek P J. J Virol. 1997;71:451–457. doi: 10.1128/jvi.71.1.451-457.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tang C K, Draper D E. Cell. 1989;57:531–536. doi: 10.1016/0092-8674(89)90123-2. [DOI] [PubMed] [Google Scholar]
- 29.Shamoo Y, Tam A, Konigsberg W H, Williams K R. J Mol Biol. 1993;232:89–104. doi: 10.1006/jmbi.1993.1372. [DOI] [PubMed] [Google Scholar]
- 30.Du Z, Hoffman D W. Nucleic Acids Res. 1997;25:1130–1135. doi: 10.1093/nar/25.6.1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tu C, Tzeng T-H, Bruenn J A. Proc Natl Acad Sci USA. 1992;89:8636–8640. doi: 10.1073/pnas.89.18.8636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Somogyi P, Jenner A J, Brierley I, Inglis S C. Mol Cell Biol. 1993;13:6931–6940. doi: 10.1128/mcb.13.11.6931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Atkins J F, Gesteland R F. Nat Struct Biol. 1999;6:206–207. doi: 10.1038/6642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miller W A, Dinesh-Kumar S P, Paul C P. Crit Rev Plant Sci. 1995;14:179–211. [Google Scholar]
- 35.Veidt I, Lot H, Leiser M, Scheidecker D, Guilley H, Richards K, Jonard G. Nucleic Acids Res. 1988;16:9917–9932. doi: 10.1093/nar/16.21.9917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Garcia A, van Duin J, Pleij C W A. Nucleic Acids Res. 1993;21:401–406. doi: 10.1093/nar/21.3.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.ten Dam E B, Verlaan P W G, Pleij C W A. RNA. 1996;1:146–154. [PMC free article] [PubMed] [Google Scholar]
- 38.Su L, Chen L, Egli M, Berger J M, Rich A. Nat Struct Biol. 1999;6:285–292. doi: 10.1038/6722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gonzalez R L, Tinoco I. J Mol Biol. 1999;289:1267–1282. doi: 10.1006/jmbi.1999.2841. [DOI] [PubMed] [Google Scholar]
- 40.Napthine S, Liphardt J, Bloys A, Routledge S, Brierley I. J Mol Biol. 1999;288:305–320. doi: 10.1006/jmbi.1999.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liphardt J, Napthine S, Kontos H, Brierley I. J Mol Biol. 1999;288:321–335. doi: 10.1006/jmbi.1999.2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kang H, Hines J V, Tinoco I. J Mol Biol. 1996;259:135–147. doi: 10.1006/jmbi.1996.0308. [DOI] [PubMed] [Google Scholar]
- 43.Du Z, Holland J A, Hansen M R, Giedroc D P, Hoffman D W. J Mol Biol. 1997;270:464–470. doi: 10.1006/jmbi.1997.1127. [DOI] [PubMed] [Google Scholar]
- 44.Wills N M, Gesteland R F, Atkins J F. EMBO J. 1994;13:4137–4144. doi: 10.1002/j.1460-2075.1994.tb06731.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shen L, Tinoco I. J Mol Biol. 1995;247:963–978. doi: 10.1006/jmbi.1995.0193. [DOI] [PubMed] [Google Scholar]
- 46.Kang H, Tinoco I. Nucleic Acids Res. 1997;25:1943–1949. doi: 10.1093/nar/25.10.1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen X, Kang H, Shen L X, Chamorro M, Varmus H E, Tinoco I. J Mol Biol. 1996;260:479–483. doi: 10.1006/jmbi.1996.0415. [DOI] [PubMed] [Google Scholar]
- 48.Alam S L, Wills N M, Ingram J A, Atkins J F, Gesteland R F. J Mol Biol. 1999;288:837–852. doi: 10.1006/jmbi.1999.2713. [DOI] [PubMed] [Google Scholar]
- 49.Horsfield J A, Wilson D N, Mannering S A, Adamski F M, Tate P T. Nucleic Acids Res. 1995;23:1487–1494. doi: 10.1093/nar/23.9.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]


