We hypothesized that the gold standard for diagnosing leptospirosis is imperfect. We used Bayesian latent class models and random-effects meta-analysis to test this hypothesis and to determine the true accuracy of a range of alternative tests for leptospirosis diagnosis.
Abstract
Background. We observed that some patients with clinical leptospirosis supported by positive results of rapid tests were negative for leptospirosis on the basis of our diagnostic gold standard, which involves isolation of Leptospira species from blood culture and/or a positive result of a microscopic agglutination test (MAT). We hypothesized that our reference standard was imperfect and used statistical modeling to investigate this hypothesis.
Methods. Data for 1652 patients with suspected leptospirosis recruited during three observational studies and one randomized control trial that described the application of culture, MAT, immunofluorescence assay (IFA), lateral flow (LF) and/or PCR targeting the 16S rRNA gene were reevaluated using Bayesian latent class models and random-effects meta-analysis.
Results. The estimated sensitivities of culture alone, MAT alone, and culture plus MAT (for which the result was considered positive if one or both tests had a positive result) were 10.5% (95% credible interval [CrI], 2.7%–27.5%), 49.8% (95% CrI, 37.6%–60.8%), and 55.5% (95% CrI, 42.9%–67.7%), respectively. These low sensitivities were present across all 4 studies. The estimated specificity of MAT alone (and of culture plus MAT) was 98.8% (95% CrI, 92.8%–100.0%). The estimated sensitivities and specificities of PCR (52.7% [95% CrI, 45.2%–60.6%] and 97.2% [95% CrI, 92.0%–99.8%], respectively), lateral flow test (85.6% [95% CrI, 77.5%–93.2%] and 96.2% [95% CrI, 87.7%–99.8%], respectively), and immunofluorescence assay (45.5% [95% CrI, 33.3%–60.9%] and 96.8% [95% CrI, 92.8%–99.8%], respectively) were considerably different from estimates in which culture plus MAT was considered a perfect gold standard test.
Conclusions. Our findings show that culture plus MAT is an imperfect gold standard against which to compare alterative tests for the diagnosis of leptospirosis. Rapid point-of-care tests for this infection would bring an important improvement in patient care, but their future evaluation will require careful consideration of the reference test(s) used and the inclusion of appropriate statistical models.
The clinical manifestations of leptospirosis range from a mild influenza-like illness to multiorgan failure and death. The common signs and symptoms of this infection fail to discriminate leptospirosis from a range of other infectious diseases that occur in the tropics, including dengue fever, malaria, and rickettsial infection [1]. Although laboratory diagnosis has the potential to guide the management of patients with leptospirosis, this is not currently achieved in most regions of the world because of a lack of point-of-care tests. Once such tests become available, however, it will be important to ensure that the diagnostic reference standard against which they are compared during clinical evaluation is robust.
We have undertaken several therapeutic and diagnostic evaluation studies involving patients presenting to hospitals in Thailand with suspected leptospirosis [2–5]. The reference tests used were a combination of culture for Leptospira species and the microscopic agglutination test (MAT). Definite leptospirosis was defined as the isolation of Leptospira species from a normally sterile site and/or a 4-fold increase in the MAT titer between acute- and convalescent-phase serum samples or a single MAT titer of ≥1:400. This is consistent with the published recommendations of the World Health Organization Leptospirosis Burden Epidemiology Reference Group [6].
We began to question the accuracy of the recommended approach after becoming increasingly aware that some patients with suspected leptospirosis who had positive results of alternative tests, such as a real-time polymerase chain reaction (PCR) targeting the gene encoding the 16S ribosomal RNA (rRNA) subunit, a lateral flow test, and/or an immunofluorescence assay (IFA), were negative for leptospirosis on the basis of our reference standard. This was a consistent finding across different studies in which we had obtained a convalescent-phase specimen from the majority of cases. We hypothesized that the reference standard was imperfect and that the accuracy of alternative diagnostic tests, estimated using the gold standard, was biased. We sought an appropriate statistical model with which to determine the true accuracy of alternative tests in this situation.
Bayesian latent class models have been increasingly used to evaluate the true accuracy of diagnostic tests and do not require the assumption that any test or combination of tests is perfect [7, 8]. The objective of this study was to use Bayesian latent class models to reanalyze individual-level data from 4 existing data sets gathered during studies of patients presenting to the hospital with suspected leptospirosis. On the basis of these findings, we estimated the true accuracy of a range of serological and molecular diagnostic tests for leptospirosis and determined the impact of an imperfect gold standard on the reported accuracies of alternative tests. Information from all studies was combined using Bayesian random-effects meta-analysis to further support the observations from individual studies.
METHODS
We followed a standard protocol for meta-analyses [9], together with the methods recommended by the Cochrane Diagnostic Test Accuracy Working Group, the STARD (Standards for the Reporting of Diagnostic Accuracy) statement, and the PRISMA (Preferred Reporting Items for Systematic Reviews and Meta-analyses) statement [10].
Search Strategy and Study Selection
Our aim was to reanalyze complete individual-level data sets created by us during hospital-based studies of suspected leptospirosis conducted in northeast Thailand between 2000 and 2010 (Table 1). All studies had undertaken prospective enrollment of adult patients (age, >14 years) with suspected leptospirosis and had used a combination of blood culture for Leptospira species and MAT (hereafter, “culture plus MAT”) as the diagnostic reference standard. Studies were selected if individual-level data sets were available for analysis. For patients included in >1 study, only data from the first study were used in the analysis.
Table 1.
Characteristics of Populations and Studies in Thailand Included in the Analyses
| Studya | Authors | Province(s) | Year | Study Design | Sample Sizeb | Diagnostic Tests |
|---|---|---|---|---|---|---|
| A | Thaipadungpanit et al [2] | Udon Thani | 2000–2001 | Prospective, observational | 371 | Culture, MAT, PCR, and LF |
| B | Wuthiekanun et al [3] | Udon Thani | 2000–2002 | Prospective, observational | 496 | Culture, MAT, IFA |
| C | Phimda et al [4] | Udon Thani, Nakorn Rachasima, Chaiyapoom, Chumphon | 2003–2005 | Multicenter, randomized controlled trial | 314 | Culture, MAT |
| D | Wuthiekanun et al [5] | Udon Thani, Maha Sarakarm, Yasothorn, Chainut, Rayong, Chanthaburi, Prachuap Khiri Khun, Phattalung | 2003–2004 | Multicenter, prospective, observational | 471 | Culture and MAT |
Abbreviations: IFA, immunofluorescence assay; LF, lateral flow test; MAT, microscopic agglutination test; PCR, polymerase chain reaction.
a Studies are ordered chronologically.
b Records of patients for whom not all of the intended tests were performed and records duplicated among studies are excluded.
Ethics Statement
Ethical approval for all studies included in the analysis was obtained from the Ministry of Public Health, Thailand, and the Oxford Tropical Research Ethics Committee, United Kingdom. Written inform consent was obtained from each subject enrolled into these studies [2–5].
Diagnostic Tests
All diagnostic tests that were used in each study were evaluated. In each study, blood was collected on the day of admission and cultured for Leptospira organisms, as described previously [3]. Serum samples (5 mL) collected on admission and, if available, at a 2-week follow-up visit were used for serological testing. Serum was stored at −80°C between collection and serological testing. The MAT was performed at the World Health Organization/United Nations Food and Agriculture Organization/World Animal Health Organization Collaborating Center for Reference and Research on Leptospirosis in Brisbane, Australia. The panel of serovars used in the MAT included representative serovars from all serogroups known to cause leptospirosis in Thailand. A real-time PCR assay targeting the 16S rRNA subunit, a lateral flow test (Leptotek, BioMerieux, the Netherlands), and an in-house IFA were performed as described previously [2, 11, 12]. The MAT detects crude antibodies against Leptospira organisms, the lateral flow test detects immunoglobulin M (IgM), and the IFA detects immunoglobulin G, immunoglobulin A, and IgM [11, 12]. All tests were performed by experienced technicians. The readers of results of culture, MAT, and other diagnostic tests in each study were blinded to the results of the other tests and any clinical information.
Statistical Analysis
Culture Plus MAT as Gold Standard Model
Five diagnostic tests (culture, MAT, IFA, lateral flow test, and PCR) were analyzed for each of the studies, using culture plus MAT as the gold standard. Prevalence, sensitivity, specificity, and positive and negative predictive values for each of the 6 tests were calculated using Stata 11.1 (Stata, College Station, TX).
Bayesian Latent Class Models
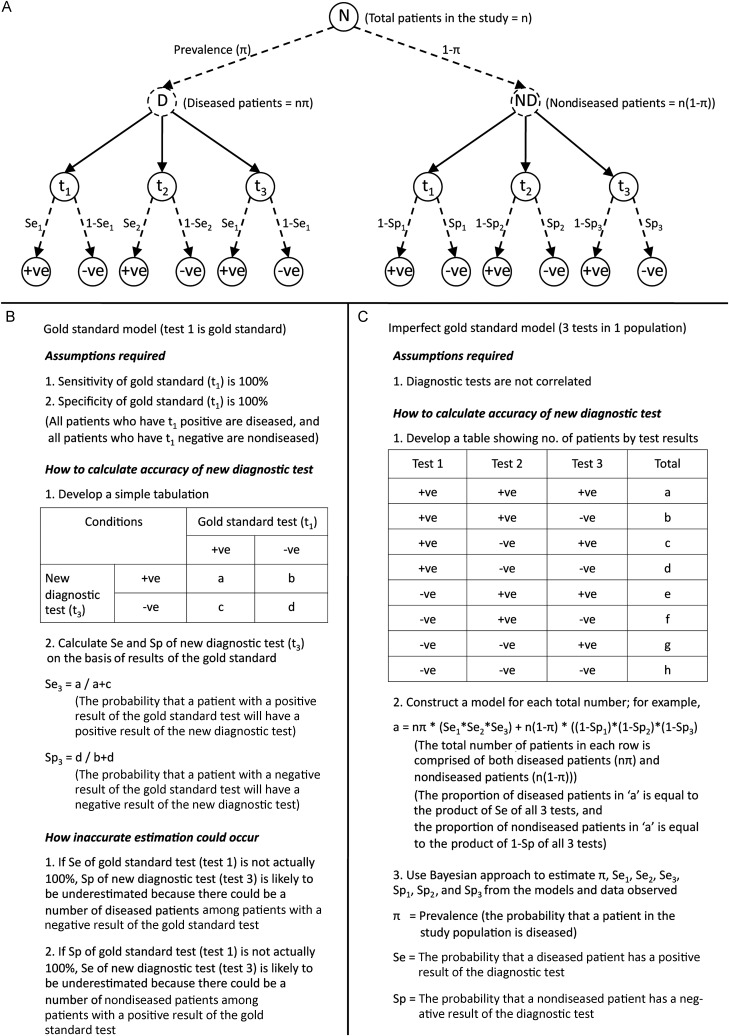
Use of latent class models and Bayesian latent class models to determine the accuracy of diagnostic tests when the accuracy of the gold standard is imperfect or unknown has been described in detail elsewhere [13–15]. Figure 1 illustrates how the imperfect gold standard model estimates unbiased accuracies of diagnostic tests in one example scenario: application of 3 diagnostic tests to 1 study population. In brief, use of Bayesian latent class models does not assume that any test or a combination of any tests is perfect but considers that each test could be imperfect in diagnosing the true disease status. The true disease status of the patient population is then defined on the basis of the overall prevalence (the probability that a patient with suspected leptospirosis is truly infected) [13–15]. Latent class models estimate the prevalence and accuracy of each test on the basis of the observed frequency of the possible combinations of test results.
Figure 1.
Schematic illustration of the use of Bayesian latent class model to obtain unbiased estimates of accuracy of diagnostic tests. A, Overview of all possible outcomes of diagnostic tests, based on true disease status, if 3 diagnostic tests are applied to 1 study population. Broken lines represent unknown parameters. Patients under evaluation could be either diseased or nondiseased, and prevalence represents the probability that a patient is diseased. Solid lines represent the application of all 3 diagnostic tests (t1, t2, and t3) to every patient in the study. Test results are conditional on the sensitivity and specificity of each test. True disease status (diseased or nondiseased) is a latent variable, as it is not directly observed but can be estimated as the prevalence in the Bayesian latent class model. B and C, Comparison of how to estimate the accuracy of diagnostic tests, using the gold standard model (B) and the imperfect gold standard model (C).
To estimate the accuracy of a diagnostic test by use of latent class models, the best-fitting model, as determined by the presence or absence of correlation between diagnostic tests in the model, should be used [14, 15]. Possible correlations we evaluated were based on existing knowledge and external evidence. Therefore, correlation among antigenic tests (culture and PCR) and correlation among serological tests (MAT, IFA, and the lateral flow test) were considered. All models assumed that no prior information (noninformative priors) about the unknown parameters (ie, prevalence, sensitivities, and specificities) was available, except that the specificity of culture was fixed at 100%. For multicenter studies [4, 5], the models also assumed that sensitivities and specificities of culture and MAT were consistent over different study sites. All parameters and associated 95% credible intervals (CrIs) were estimated using WinBUGS 1.4 [16].
To estimate the overall accuracy of culture alone, MAT alone, and culture plus MAT across all studies, the information obtained from the best-fitting Bayesian latent class model for each data set was combined using a Bayesian meta-analysis model [17]. Random effects were used to account for differences in the sensitivities of culture and MAT and the specificity of MAT between studies. The ranges of each parameter from the Bayesian latent class model with the best fit obtained for individual data sets were used as informative priors in the meta-analysis model [17]. Appendixes 1 and 2 (Supplementary Materials) provide full data sets and all of the models used, respectively. Appendix 3 (Supplementary Materials) provides details about the method and the results for the best-fitting model selection.
Sensitivity Analyses
Sensitivity analyses were performed in which patients without convalescent-phase samples were excluded and also in which different prior information were used [18].
RESULTS
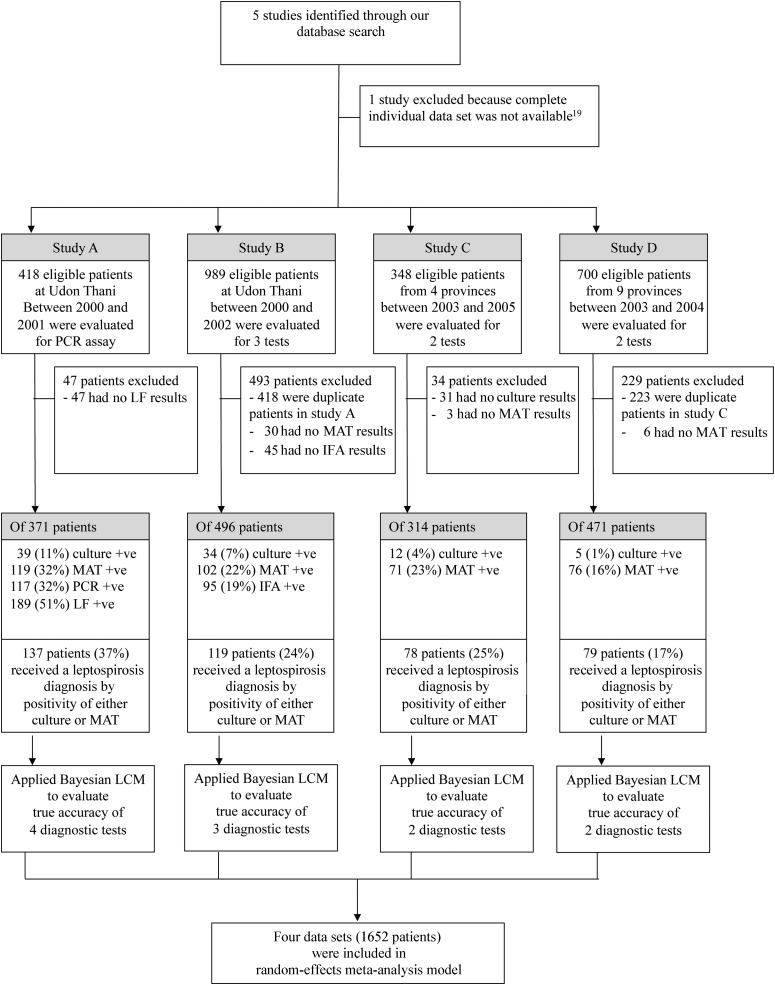
The flow diagram in Figure 2 provides an overview of the study. Five studies conducted by us between 2000 and 2010 that used or evaluated diagnostic tests to investigate patients presenting to the hospital with suspected leptospirosis were searched for the presence of complete individual-level data. One study was excluded because individual test results were not available [19]. Of 2455 records from the 4 remaining data sets, 803 were excluded because they were duplicated in ≥1 of the other studies (n = 641) or because not all of the intended tests were undertaken (n = 162). Therefore, 4 data sets involving 1652 patients were included in the analysis (Table 1) [2–5]. Studies A and B were prospective observational studies conducted at a single hospital in Udon Thani, in northeast Thailand, in which culture, MAT, and at least 1 additional test (PCR, lateral flow test, or IFA) was undertaken in all cases. Studies C and D were prospective multicenter studies in which culture and MAT were undertaken in all cases. All serological tests had been performed using paired samples, when available. The median duration of illness prior to admission was 4 days, and the median duration of illness prior to obtaining the convalescent-phase serum sample was 14 days.
Figure 2.
Study flow diagram.
We first assumed that the combination of culture plus MAT was a perfect reference test (100% sensitivity and 100% specificity). This gave an estimated prevalence for leptospirosis of 36.9%, 24.0%, 24.8%, and 16.8% for studies A, B, C, and D, respectively. The sensitivity of culture alone was low and varied significantly among studies (28.5%, 28.6%, 15.4%, and 6.3% for studies A, B, C, and D, respectively). The sensitivity of MAT was high but also varied significantly among studies (86.9%, 85.7%, 91.0%, and 96.2% for studies A, B, C, and D, respectively). PCR, the lateral flow test, and IFA were found to have either low sensitivity (55.5%, 87.6%, and 64.7%, respectively) or low specificity (82.5%, 70.5%, and 95.2%, respectively).
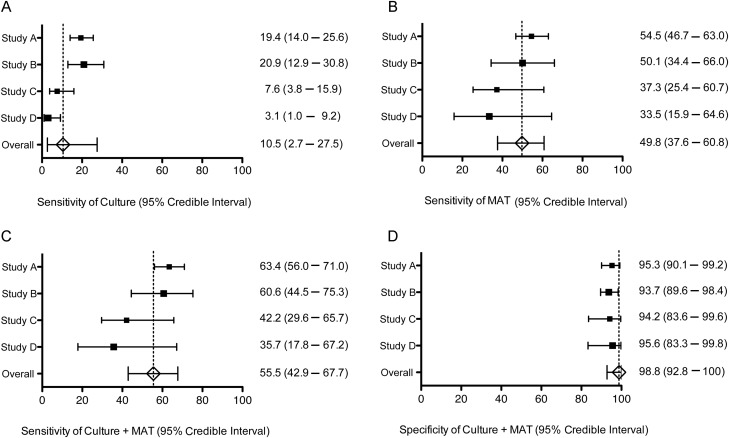
Bayesian latent class models were then used to obtain an estimate of the accuracy of each diagnostic test, without the assumption that the reference test was perfect. In the first stage, we defined the best-fitting Bayesian latent class model for each data set by determining the presence of correlations between antigenic tests (culture and PCR) and serological tests (MAT, IFA, and the lateral flow test) (Appendix 3). The best-fitting model for study A was the model that included correlations between culture and PCR and between MAT and the lateral flow test. The best-fitting model for study B was the model that included correlation between MAT and IFA. The model without correlation between diagnostic tests was selected for studies C and D because we did not expect a correlation between culture and MAT. Sensitivities and specificities of culture, MAT, and culture plus MAT, estimated by Bayesian latent class model for each study, are shown in Figure 3. These findings formed the basis for choosing the model with correlation between culture and PCR, between MAT and the lateral flow test, and between MAT and IFA for the meta-analysis.
Figure 3.
Forest plot of sensitivity and specificity of culture (A), microagglutination test (MAT; B), and the combination of culture and MAT (culture + MAT; C and D) for leptospirosis estimated by Bayesian latent class models (LCMs) and random-effects meta-analysis models. Squares represent median estimates of the sensitivities and the specificities, and the size of the square represents the size of the study. Horizontal lines represent 95% credible intervals of the estimates. Bayesian LCM assuming conditional dependence between culture and PCR and between MAT and lateral flow (LF) was used for study A, and Bayesian LCM assuming conditional dependence between MAT and immunofluorescence assay was used for study B. Bayesian LCM assuming conditional independence between tests and consistency of test accuracies between study sites was used for studies C and D. Meta-analysis was performed by application of random-effects variables into the combined data set of all 4 studies, assuming conditional dependence between culture and PCR, between MAT and LF, and between MAT and IFA. Abbreviations: IFA, immunofluorescence assay; LCM, latent class model; LF, lateral flow; MAT, microagglutination test; PCR, polmerase chain reaction.
Data across all 4 studies were then combined and analyzed using a Bayesian latent class random-effects meta-analysis model, which demonstrated that all of the estimated parameters were considerably different from those estimated when culture plus MAT was assumed to be perfect. The meta-analysis model indicated that culture, MAT, and culture plus MAT had very low sensitivities of 10.5% (95% CrI, 2.7%–27.5%), 49.8% (95% CrI, 37.6%–60.8%), and 55.5% (95% CrI, 42.9%–67.7%), respectively (Figure 3 and Table 2). The specificity of both MAT alone and of culture plus MAT was 98.8% (95% CrI, 92.8%–100%). This means that the prevalence of leptospirosis estimated in each data set using the model was much higher than if relying on culture plus MAT (eg, 57.4% vs 36.9% for study A). Of the 2 antigenic tests, PCR had the highest sensitivity, at 52.7% (95% CrI, 45.2%–60.6%). Among the 3 serological tests, the lateral flow test had the highest sensitivity, at 85.6% (95% CrI, 77.5%–93.2%). Because it is possible that a combination of PCR and a serological test could be used as point-of-care diagnostic tests for leptospirosis, using positive results of either test, the sensitivity and specificity of PCR plus the lateral flow test were calculated by the model; these were 93.2% (95% CrI, 88.8%–96.9%) and 93.1% (95% CrI, 84.1%–98.5%), respectively (Table 2).
Table 2.
Prevalence of Leptospirosis and Accuracy of Diagnostic Tests, Determined Using Culture Plus Microagglutination Tests as the Gold Standard or Bayesian Latent Class and Random-Effects Meta-analysis Models
| Parameters | Culture Plus MAT as Gold Standard (95% CI)a | Bayesian Model (95% CrI)b |
|---|---|---|
| Prevalence | ||
| Data set A | 36.9 (32.0–41.9) | 57.4 (49.8–64.3) |
| Data set B | 24.0 (20.3–28.0) | 38.1 (27.4–52.2) |
| Data set C | 24.8 (20.2–30.0) | 45.9 (34.4–55.5) |
| Data set D | 16.8 (13.5–20.5) | 32.6 (21.5–46.9) |
| Culture | ||
| Sensitivity | 28.5, 28.6, 15.4, and 6.3* | 10.5 (2.7–27.5) |
| Specificity | 100 | 100 |
| PPV | 100 | 100 |
| NPV | 70.5, 81.6, 78.2, and 84.1* | 45.6 (37.8–54.7) |
| MAT | ||
| Sensitivity | 86.9, 85.7, 91.0, and 96.2* | 49.8 (37.6–60.8) |
| Specificity | 100 | 98.8 (92.8–100) |
| PPV | 100 | 98.3 (88.4–100) |
| NPV | 92.9, 95.7, 97.1, and 99.2* | 59.2 (49.8–68.4) |
| Culture plus MATc | ||
| Sensitivity | 100 | 55.5 (42.9–67.7) |
| Specificity | 100 | 98.8 (92.8–100) |
| PPV | 100 | 98.5 (89.6–100) |
| NPV | 100 | 62.0 (52.1–72.2) |
| PCRd | ||
| Sensitivity | 55.5 (46.7–64.0) | 52.7 (45.2–60.6) |
| Specificity | 82.5 (77.0–87.1) | 97.2 (92.0–99.8) |
| PPV | 65.0 (56.2–73.7) | 96.2 (88.5–99.8) |
| NPV | 76.0 (70.7–81.3) | 60.4 (51.5–69.2) |
| LFd | ||
| Sensitivity | 87.6 (80.9–92.6) | 85.6 (77.5–93.2) |
| Specificity | 70.5 (64.2–76.3) | 96.2 (87.7–99.8) |
| PPV | 63.5 (56.6–70.4) | 96.9 (88.9–99.9) |
| NPV | 90.7 (86.4–94.9) | 83.3 (72.2–92.5) |
| PCR plus LFc,d | ||
| Sensitivity | 92.7 (87.0–96.4) | 93.2 (88.8–96.9) |
| Specificity | 66.2 (59.8–72.3) | 93.1 (84.1–98.5) |
| PPV | 61.7 (55.0–68.3) | 94.9 (86.8–99.0) |
| NPV | 93.9 (90.3–97.6) | 91.1 (83.4–96.2) |
| IFAd | ||
| Sensitivity | 64.7 (55.4–73.2) | 45.5 (33.3–60.9) |
| Specificity | 95.2 (92.5–97.1) | 96.8 (92.8–99.8) |
| PPV | 81.1 (71.7–88.4) | 95.2 (87.8–99.7) |
| NPV | 89.5 (86.1–92.3) | 57.1 (47.3–68.2) |
Abbreviations: CI, confidence interval; CrI, credible interval; IFA, immunofluorescence assay; LF, lateral flow test; MAT, microscopic agglutination test; NPV, negative predictive value; PCR, polymerase chain reaction; PPV, positive predictive value.
*For studies A, B, C, and D, respectively.
a Values were estimated on the basis of the observed proportion, which was determined by assuming that culture plus MAT is the gold standard (ie, 100% sensitive and 100% specific). The 95% confidence intervals were obtained using Stata 11.1 (Stata).
b Values were estimated using Bayesian latent class and random-effects meta-analysis models, assuming that culture plus MAT is imperfect. Posterior estimates and 95% credible intervals of each parameter were obtained in WinBUGs from 10 000 iterations of each of 2 chains, starting from different initial values following a burn-in period of 5000 iterations.
c Positive results of one or both tests is diagnostic for leptospirosis infection.
d Values calculated by assuming that culture plus MAT is the gold standard were based on data from studies A and B, whereas values calculated by the Bayesian model were estimated from a meta-analysis model, using the data set for all 4 studies combined.
Sensitivity Analysis
Sensitivity analysis was performed in which 555 of 1652 patients (33.6%) without convalescent-phase samples were excluded. By use of a random-effects meta-analysis model, the sensitivities of MAT, the lateral flow test, and IFA were estimated to be 70.3% (95% CrI, 44.1%–91.5%), 89.7% (95% CrI, 80.3%–97.2%), and 71.2% (95% CrI, 52.4%–89.8%), respectively, for patients with leptospirosis who had a convalescent-phase sample. The accuracies of other diagnostic tests were not substantially different from the above values, although all CrIs were wider as a consequence of the reduced information in the sample sizes. There was no substantial change when different prior information was used (Appendix 4 [Supplementary Data]).
DISCUSSION
The key findings of this study are that the true sensitivities of culture, MAT, and culture plus MAT are low. The finding that culture has a low sensitivity is neither novel nor surprising, since Leptospira organisms are only present in the blood during the first week of untreated infection, and isolation of this bacterium from clinical samples is technically demanding. Furthermore, our patient population may have consumed over-the-counter antibiotics prior to hospital presentation, which may have resulted in false-negative culture results. The proportion of patients who had taken antibiotics by the time of presentation in our studies is not known, but a study of patients presenting to a hospital in neighboring Laos showed that 57% of patients who were admitted to Mahosot Hospital and underwent investigations, including lumbar puncture, had antimicrobial activity detected in their urine [20]. Other possible explanations for the low sensitivity of culture are that viable Leptospira species are difficult to recover from clinical samples, using the existing culture methods. Of note, cultures were maintained for at least 6 months before results were deemed to be negative. More worrying is the low sensitivity of MAT, since this is central to the case definition of definite leptospirosis and is widely used.
There are several potential reasons why MAT had low sensitivity in our setting. Antibodies to Leptospira may take several weeks to become detectable by MAT [21]. In the studies described here, the second (ie, convalescent-phase) serum sample was collected at least 10 days after the start of symptoms that were attributed to leptospirosis, but it is possible that this interval was too short in some cases [22]. In common with other research studies and reflecting real life, we also failed to obtain a convalescent-phase serum specimen from 34% of patients, either because they died or because they were discharged and were lost to follow-up. The results from our sensitivity analysis also show that the sensitivity of MAT was 70.3% in the ideal situation, in which convalescent-phase samples were obtained from all patients. This increase in sensitivity is consistent with existing knowledge and is comparable to a previous estimate (76%), in which only patients with culture-confirmed leptospirosis were considered [22]. However, this also suggests that a number of patients with leptospirosis have a false-negative test result by MAT even if a convalescent-phase sample is available. Other possible explanations for the low sensitivity of MAT are that lipopolysaccharide from different Leptospira serovars induces a variable level of immune response and that the Leptospira serovars used in the MAT did not include 1 or more locally important strains, although we have no evidence that this is the case. The poor sensitivity of culture plus MAT in the real clinical setting, as shown by Bayesian latent class modeling, suggests that improvement of both tests or development of a new gold standard test is required.
Our data supported a positive correlation between both antigenic tests (ie, culture and PCR), a finding that could be interpreted as meaning that results of both culture and PCR are more likely to be positive if the burden of Leptospira organisms in blood is high and to be negative if the burden is low [2, 23]. A positive correlation was also found between serological tests (ie, between MAT and IFA and between MAT and the lateral flow test). These findings are consistent with existing knowledge.
A major effect of poor sensitivity of the reference test is that the prevalence of leptospirosis is underestimated. The estimated prevalence of leptospirosis for each study separately, as determined by Bayesian latent class modeling (32.6%–57.4%), was around double that of previous estimates that used culture plus MAT as the gold standard (16.8%–36.9%). This is credible, since 50% of our study patients with suspected leptospirosis left the hospital without a definite diagnosis following a test panel that included bacterial culture; other serological tests, including those for rickettsial infections (which are also common in our setting); radiological tests; and detailed clinical evaluation [2–5]. Our study suggests that leptospirosis may have been the cause of fever in a proportion of these cases.
Evaluation of diagnostic tests when the accuracy of the gold standard is unknown is an active area of biostatistical research, since the use of an imperfect gold standard to evaluate the accuracy and clinical usefulness of an alternative test is flawed and leads to biased results [8, 14, 17, 24]. Our study has demonstrated that culture plus MAT represents a relatively poor gold standard against which to compare alternative diagnostic tests for leptospirosis and has shown the usefulness of statistical models under such circumstances. For example, PCR had a sensitivity and specificity of 55.5% and 82.5%, respectively, when compared with culture plus MAT. When recalculated using Bayesian latent class modeling, the sensitivity and specificity of PCR were 52.7% and 97.2%, respectively, representing a test with a high degree of specificity.
Our study had several strengths, including the use of large and individual-level data sets rather than summary estimates from the published literature. Disparity of study characteristics and risk of bias were comparatively low, since all studies were conducted prospectively and by the same research unit. None of the studies were supported by diagnostic companies. In addition, we used a random-effects model, a rigorous statistical method that has been recommended by the Cochrane Diagnostic Test Accuracy Working Group as the method of choice for diagnostic meta-analyses [10].
This study also has several limitations. Use of basic Bayesian latent class models to estimate the sensitivity and specificity of each test in a population does not allow us to determine the effects that symptom duration, antimicrobials received prior to presentation, and timing of convalescent-phase samples at the level of individual patients have on these parameters. These effects could be evaluated in advanced Bayesian latent class models [25]. Correlation between IFA and the lateral flow test could not be evaluated because these tests were not performed together in any study included in the analysis. The small number of studies included also meant that important study characteristics, including differences in the prevalence of leptospirosis, and the level of reproducibility between studies of the finding of high accuracy for PCR and the lateral flow test were not assessed. The commercial lateral flow test evaluated here had a published specificity that was underestimated, compared with the gold standard, but it is not currently available [26, 27]. The currently available rapid serological tests for leptospirosis include a latex agglutination test and an IgM enzyme-linked immunosorbant assay [28–30], neither of which has been evaluated by us to date.
We conclude that our current reference testing strategy is imperfect. As a result, both the prevalence of leptospirosis in northeast Thailand and the accuracy of alternative diagnostic tests have been underestimated. There is an urgent need for rapid serological tests for leptospirosis. Our findings support the use of latent class models to evaluate such a new test against an imperfect gold standard.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online (http://cid.oxfordjournals.org). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Acknowledgments. We gratefully acknowledge the support provided by staff at the Mahidol-Oxford Tropical Medicine Research Unit and participating hospitals. The lateral flow test was provided during 2000 at no cost by BioMerieux (The Netherlands).
Disclaimer. BioMerieux had no involvement in any part of the work described, nor the writing of the manuscript.
Financial support. This study was supported by the Wellcome Trust. D. L. is supported by a project grant awarded by the Wellcome Trust (090219/Z/09/Z). S. J. P. is supported by the National Institute for Health Research Cambridge Biomedical Research Centre.
Potential conflict of interest. All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Suttinont C, Losuwanaluk K, Niwatayakul K, et al. Causes of acute, undifferentiated, febrile illness in rural Thailand: results of a prospective observational study. Ann Trop Med Parasitol. 2006;100:363–70. doi: 10.1179/136485906X112158. [DOI] [PubMed] [Google Scholar]
- 2.Thaipadungpanit J, Chierakul W, Wuthiekanun V, et al. Diagnostic accuracy of real-time PCR assays targeting 16S rRNA and lipl32 genes for human leptospirosis in Thailand: a case-control study. PLoS One. 2011;6:e16236. doi: 10.1371/journal.pone.0016236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wuthiekanun V, Chierakul W, Limmathurotsakul D, et al. Optimization of culture of Leptospira from humans with leptospirosis. J Clin Microbiol. 2007;45:1363–5. doi: 10.1128/JCM.02430-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Phimda K, Hoontrakul S, Suttinont C, et al. Doxycycline versus azithromycin for treatment of leptospirosis and scrub typhus. Antimicrob Agents Chemother. 2007;51:3259–63. doi: 10.1128/AAC.00508-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wuthiekanun V, Sirisukkarn N, Daengsupa P, et al. Clinical diagnosis and geographic distribution of leptospirosis, Thailand. Emerg Infect Dis. 2007;13:124–6. doi: 10.3201/eid1301.060718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.World Health Organization (WHO) Geneva: WHO; 2010. Report of the first meeting of leptospirosis burden epidemiology reference group; pp. 1–34. [Google Scholar]
- 7.Speybroeck N, Praet N, Claes F, et al. True versus apparent malaria infection prevalence: the contribution of a Bayesian approach. PLoS One. 2011;6:e16705. doi: 10.1371/journal.pone.0016705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Limmathurotsakul D, Jamsen K, Arayawichanont A, et al. Defining the true sensitivity of culture for the diagnosis of melioidosis using Bayesian latent class models. PLoS One. 2010;5:e12485. doi: 10.1371/journal.pone.0012485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deeks JJ. Systematic reviews in health care: systematic reviews of evaluations of diagnostic and screening tests. BMJ. 2001;323:157–62. doi: 10.1136/bmj.323.7305.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leeflang MM, Deeks JJ, Gatsonis C, Bossuyt PM. Systematic reviews of diagnostic test accuracy. Ann Intern Med. 2008;149:889–97. doi: 10.7326/0003-4819-149-12-200812160-00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smits HL, Eapen CK, Sugathan S, et al. Lateral-flow assay for rapid serodiagnosis of human leptospirosis. Clin Diagn Lab Immunol. 2001;8:166–9. doi: 10.1128/CDLI.8.1.166-169.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Appassakij H, Silpapojakul K, Wansit R, Woodtayakorn J. Evaluation of the immunofluorescent antibody test for the diagnosis of human leptospirosis. Am J Trop Med Hyg. 1995;52:340–3. doi: 10.4269/ajtmh.1995.52.340. [DOI] [PubMed] [Google Scholar]
- 13.Zhou XM, Mcclish DK, Obuchowski NA. Methods for correcting imperfect standard bias. In: Statistical methods in diagnostic medicine. 1st ed. New York: Wiley-Interscience; 2002. p. 40. [Google Scholar]
- 14.Joseph L, Gyorkos TW, Coupal L. Bayesian estimation of disease prevalence and the parameters of diagnostic tests in the absence of a gold standard. Am J Epidemiol. 1995;141:263–72. doi: 10.1093/oxfordjournals.aje.a117428. [DOI] [PubMed] [Google Scholar]
- 15.Dendukuri N, Joseph L. Bayesian approaches to modeling the conditional dependence between multiple diagnostic tests. Biometrics. 2001;57:158–67. doi: 10.1111/j.0006-341x.2001.00158.x. [DOI] [PubMed] [Google Scholar]
- 16.Lunn D, Spiegelhalter D, Thomas A, Best N. The BUGS project: evolution, critique and future directions. Stat Med. 2009;28:3049–67. doi: 10.1002/sim.3680. [DOI] [PubMed] [Google Scholar]
- 17.Chu H, Chen S, Louis TA. Random effects models in a meta-analysis of the accuracy of two diagnostic tests without a gold standard. J Am Stat Assoc. 2009;104:512–23. doi: 10.1198/jasa.2009.0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spiegelhalter D, Abrams K, Myles J. Bayesian approaches to clinical trials and health-care evaluation. West Sussex, United Kingdom: Wiley; 2004. [Google Scholar]
- 19.Suputtamongkol Y, Niwattayakul K, Suttinont C, et al. An open, randomized, controlled trial of penicillin, doxycycline, and cefotaxime for patients with severe leptospirosis. Clin Infect Dis. 2004;39:1417–24. doi: 10.1086/425001. [DOI] [PubMed] [Google Scholar]
- 20.Moore CE, Sengduangphachanh A, Thaojaikong T, et al. Enhanced determination of Streptococcus pneumoniae serotypes associated with invasive disease in Laos by using a real-time polymerase chain reaction serotyping assay with cerebrospinal fluid. Am J Trop Med Hyg. 2010;83:451–7. doi: 10.4269/ajtmh.2010.10-0225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Faine S. Guidelines for the control of leptospirosis. Geneva: WHO offset publication no 67.; 1982. [PubMed] [Google Scholar]
- 22.Cumberland P, Everard CO, Levett PN. Assessment of the efficacy of an IgM-ELISA and microscopic agglutination test (MAT) in the diagnosis of acute leptospirosis. Am J Trop Med Hyg. 1999;61:731–4. doi: 10.4269/ajtmh.1999.61.731. [DOI] [PubMed] [Google Scholar]
- 23.Ahmed A, Engelberts MF, Boer KR, Ahmed N, Hartskeerl RA. Development and validation of a real-time PCR for detection of pathogenic Leptospira species in clinical materials. PLoS One. 2009;4:e7093. doi: 10.1371/journal.pone.0007093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Banoo S, Bell D, Bossuyt P, et al. Evaluation of diagnostic tests for infectious diseases: general principles. Nat Rev Microbiol. 2006;4:S20–32. doi: 10.1038/nrmicro1570. [DOI] [PubMed] [Google Scholar]
- 25.Bernatsky S, Lix L, Hanly JG, et al. Surveillance of systemic autoimmune rheumatic diseases using administrative data. Rheumatol Int. 2011;31:549–54. doi: 10.1007/s00296-010-1591-2. [DOI] [PubMed] [Google Scholar]
- 26.Wagenaar JF, Falke TH, Nam NV, et al. Rapid serological assays for leptospirosis are of limited value in southern Vietnam. Ann Trop Med Parasitol. 2004;98:843–50. doi: 10.1179/000349804X3207. [DOI] [PubMed] [Google Scholar]
- 27.Sehgal SC, Vijayachari P, Sugunan AP, Umapathi T. Field application of Lepto lateral flow for rapid diagnosis of leptospirosis. J Med Microbiol. 2003;52:897–901. doi: 10.1099/jmm.0.05064-0. [DOI] [PubMed] [Google Scholar]
- 28.Smits HL, Chee HD, Eapen CK, et al. Latex based, rapid and easy assay for human leptospirosis in a single test format. Trop Med Int Health. 2001;6:114–8. doi: 10.1046/j.1365-3156.2001.00675.x. [DOI] [PubMed] [Google Scholar]
- 29.Obregon AM, Fernandez C, Rodriguez I, Balbis Y, Martinez B, Rodriguez J. Latex agglutination system for the rapid diagnosis of leptospirosis in Cuba. Rev Panam Salud Publica. 2004;16:259–65. doi: 10.1590/s1020-49892004001000005. [DOI] [PubMed] [Google Scholar]
- 30.Bajani MD, Ashford DA, Bragg SL, et al. Evaluation of four commercially available rapid serologic tests for diagnosis of leptospirosis. J Clin Microbiol. 2003;41:803–9. doi: 10.1128/JCM.41.2.803-809.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.