Abstract
Conformational changes in the side chains are essential for protein-protein binding. Rotameric states and unbound-to-bound conformational changes in the surface residues were systematically studied on a representative set of protein complexes. The side-chain conformations were mapped onto dihedral angles space. The variable threshold algorithm was developed to cluster the dihedral angle distributions and to derive rotamers, defined as the most probable conformation in a cluster. Six rotamer libraries were generated: full surface, surface non-interface, and surface interface - each for bound and unbound states. The libraries were used to calculate the probabilities of the rotamer transitions upon binding. The stability of amino acids was quantified based on the transition maps. The non-interface residues stability was higher than that of the interface. Long side chains with three or four dihedral angles were less stable than the shorter ones. The transitions between the rotamers at the interface occurred more frequently than on the non-interface surface. Most side chains changed conformation within the same rotamer or moved to an adjacent rotamer. The highest percentage of the transitions was observed primarily between the two most occupied rotamers. The probability of the transition between rotamers increased with the decrease of the rotamer stability. The analysis revealed characteristics of the surface side-chain conformational transitions that can be utilized in flexible docking protocols.
Keywords: conformational transition, induced fit, protein-protein interactions, protein docking, molecular recognition
INTRODUCTION
The knowledge of protein-protein interactions is important for understanding protein function. The rapidly increasing amount of experimentally determined structures of proteins and protein-protein complexes provides foundation for research on protein interactions and complex formation. Protein interfaces are often described by their size, shape, amino acid composition, and a variety of other structural and physicochemical characteristics.1–4 Structural changes in proteins upon complex formation are the subject of many studies.5–14 Different models have been proposed for the binding process including an early concept of “lock and key,”15 induced-fit,16 and conformational selection.8–10,14,17,18
An earlier study by Betts and Sternberg19 described flexibility of the side chains and the backbone in proteins upon binding, without pointing the specific directions of the changes. The conclusion was that the interface conformational change is larger than that of the non-interface. The side-chain flexibility in small ligand-receptor binding was studied by Najmanovich et al.20 The side-chain flexibility analysis in the first two dihedral angles of paired unbound proteins21 determined that buried residues were inflexible, having similar conformations in different crystal structures. Ile, Thr, Asn, Asp, and the large aromatics showed limited flexibility when exposed on the protein surface, whereas Ser, Lys, Arg, Met, Gln, and Glu were found to be flexible. Directions of the side-chain conformational changes were studied by Koch et al.22 for five amino acid types. Beglov et al.23 found that the end group positions change < 1Å upon association for > 60% of the interface side chains. The study also determined that often an interface side-chain conformation in the bound state can be selected from a small ensemble of low energy rotamers and the corresponding side-chain conformation in the unbound state.
Guharoy et al.24 studied the unbound-to-bound rotamer transitions in the first two dihedral angles considered separately from each other and showed that the interface residues undergo larger conformational changes than the other surface residues, and the larger flexibility is associated with longer side chains. Three states (g−, t, g+) defined by Dunbrack and Cohen25 were used for each of the dihedral angles. The results showed that often an inter-rotamer transition occurs in the direction of a more occupied state.
This study expands our previous analysis of side-chain conformational changes upon protein binding26 where the extent and frequency of conformational transition were calculated for all dihedral angles. We showed that the scale of the conformational changes increases from the near backbone dihedral angle to the most distant one, for most amino acid residues. The opposite trend was found in the residues with symmetric aromatic (Phe and Tyr) and charged (Asp and Glu) groups, where the first dihedral angle, closest to the backbone, changes most. In general, short and long side chains were shown to have different propensities for conformational change, in agreement with the results by Guharoy et al.24 Long side chains with three or more dihedral angles are often subject to large conformational transition. Shorter residues with one or two dihedral angles typically undergo local conformational changes not leading to a conformational transition.
In the current study, we developed the variable threshold algorithm to cluster the dihedral angle distributions and to derive the most probable side-chain conformations in the clusters that define rotamers. We compiled interface, non-interface, and full surface rotamer libraries of amino acids in bound and unbound proteins, considering all dihedral angles of a particular amino acid simultaneously. To generate the libraries, we used a non-redundant set of protein-protein complexes and corresponding unbound structures from Dockground.27 The libraries were used to calculate the probabilities and the percentages of unbound-to-bound inter- and intra-rotamer transitions of interface and non-interface residues and to analyze stability of the rotamers for all amino acid types. The results point to important conformational characteristics of protein binding and provide guidelines for docking methodologies.
METHODS
The analysis was performed on the non-redundant Dockground docking benchmark set 3,27 which contains bound and corresponding unbound protein X-ray structures. The dataset consists of 233 complexes, with the unbound structures of both interacting proteins for 99 complexes, and the unbound structure of one interacting protein for 134 complexes. The dataset has sequence identity between bound and unbound structures in a complex > 97%, sequence identity between complexes < 30%, and excludes homomultimers, crystal packing, and obligate interactions.
The analysis was restricted to the surface residues, with an assumption that they play the major role in binding. The surface residues were defined as those having relative solvent-accessible surface area (RASA) ≥ 25% in bound and unbound state (the bound state of a protein was considered without its interaction partner in complex). The interface residues were defined as the surface residues, which lose > 1 Å2 solvent-accessible surface area (SASA) upon binding, calculated by NACCESS.28 The residue statistics is summarized in Table S1.
The side chain conformation was represented by dihedral angles, calculated by Dang.29 The angles varied in (−180°, 180°) interval, with the exception of the last χ angle in Phe, Tyr, Asp and Glu, which due to the symmetry of the aromatic and charged groups were reduced30 to Asp χ2 (−90°, 90°), Glu χ3 (−90°, 90°), Phe χ2 (−30°, 150°), and Tyr χ2 (−30°, 150°).
Clustering in the dihedral space: Variable Threshold algorithm
The dihedral angles distributions were calculated for each residue type in bound and unbound structures in the multidimensional space of all dihedral angles. To examine different aspects of the side chains conformations, the distributions were clustered by a novel Variable Threshold (VT) algorithm, which is a hierarchical generalization of the QT clustering31 algorithm, and is more applicable to elongated multidimensional samplings. The original QT clustering is a method of partitioning data, designed for gene clustering. It constructs disjoint clusters with the maximum occupancy, including points close to the cluster, until the diameter of the cluster surpasses the threshold. The dihedral angles distributions have few distinctive local peaks, but their vicinity has no regular shape, and is usually elongated along the last angle. Thus, to map the non-regular areas by clusters, we implemented the multi-stage cluster expansion algorithm. In the algorithm, the clusters expand into the non-regular dense areas with a decreasing clustering radius, which depends on VT step i as Ri+1 = Ri/2. The initial radii R0 given in Table I were optimized for each residue to maximize the coverage of the dihedral angles distribution functions (Figure 1). At the first stage, each point of the distribution is considered as a potential origin of a cluster with radius R0. The most populated candidate sphere is selected and marked as the first cluster. Spheres that overlap with the selected cluster are removed from consideration and the entire procedure is repeated on a smaller set of points that does not include points in the first cluster. A predefined number of large non-overlapping clusters (the procedure to define the cluster number is described in “Optimization of the initial parameters” below) is generated.
Table I.
Initial clustering radii.
| Amino acid | Radius, deg |
|---|---|
| Ser | 35 |
| Val | 35 |
| Thr | 35 |
| Cys | 35 |
| Pro | 35 |
| Ile | 35 |
| Leu | 35 |
| Asn | 55 |
| Asp | 35 |
| His | 35 |
| Phe | 35 |
| Tyr | 35 |
| Trp | 35 |
| Gln | 50 |
| Glu | [50, 50, 40]a |
| Met | 45 |
| Lys | 50 |
| Arg | 50 |
Ellipsoid with radii [50, 50, 40] was used instead of a sphere
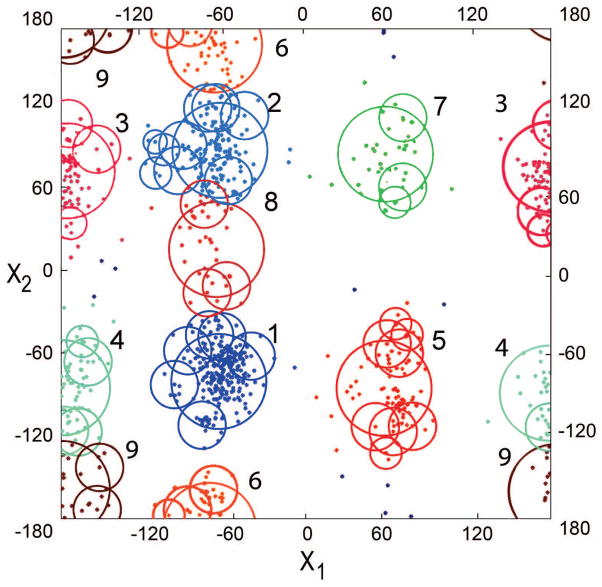
Figure 1. Clustering of Histidine conformations in dihedral space.
Numbers correspond to the clusters rank. Points are colored according to clusters. Due to the dihedral space periodicity, clusters 3, 4, 6 and 9 are visually separated.
At the second stage, smaller clustering spheres with a radius R1 = R0/2 are grown from within the previously defined clusters to expand the clusters. A candidate sphere is defined with the center at each point that lies within the parent spheres only. The occupancy of the candidate sphere is calculated outside the defined spheres (overlapping parts are not considered). The most populated candidate sphere (the cluster extension) is selected and added to the parent cluster, from which it was drawn. Then, the algorithm generates new candidate spheres from within the previously defined spheres (including the just added extension) and compares occupancies of the non-overlapping areas. The radius of any candidate sphere is always equal to the half radius of its parent sphere. The procedure of cluster growing is repeated while there are candidate spheres with non-zero occupancy. The smaller spheres from the second step and later may overlap. In this case, the points at the intersection are assigned to the sphere that was drawn first. The initial clusters grow concurrently at the second stage (e.g., at some VT step, one of the clusters adds a sphere of radius R0/4, and then the other cluster adds a sphere of radius R0/16). At the final step, spheres with radius two times larger than the original one are drawn from the initial spheres’ centers to cover the remaining unassigned points. Points at the intersection of spheres are assigned to the cluster with a more populated initial sphere.
A rotamer was assigned to each cluster as the most probable point defined by the largest number of cluster points that belong to a sphere of 10° radius with the center at this point. Thus, the rotamer not necessarily coincided with the center of the corresponding initial sphere. Tables S2 and S3 show surface interface and non-interface rotamer libraries in the decreasing order of occupancy of the initial sphere of the cluster. Figure 1 shows the dihedral angles distribution in all surface Histidines in the bound state.
Optimization of the initial parameters
The clustering has two input parameters: the number of the initial spheres n and the initial clustering radii. The parameters were independently defined for each amino acid by a combination of visual inspection of the dihedral angles distributions, their one-dimensional projections, and optimization of the cluster occupancy (coverage). The parameters were varied to increase the coverage, while avoiding the inclusion of any of the distribution main peaks into different clustering spheres.
Table I shows the initial sphere radii R0 for different amino acids. Small amino acids, with the exception of Asn, have R0 =35°. The distribution of Asn dihedral angles is elongated at the second angle. The average standard deviation in the rotamer clusters vary between 11° and 26° for all amino acids except Asn. The average standard deviation in the Asn rotamer clusters is 36°. Thus, a larger 55° radius was chosen for the clustering of Asn distributions. Long side chains with three or four dihedral angles had more scattered distributions than the shorter ones. Thus, larger clustering radii were used for the long chains. To optimize the number of the clusters and the initial clustering radii, the coverage (percentage of points assigned to the clusters) was maximized for the long side chains.
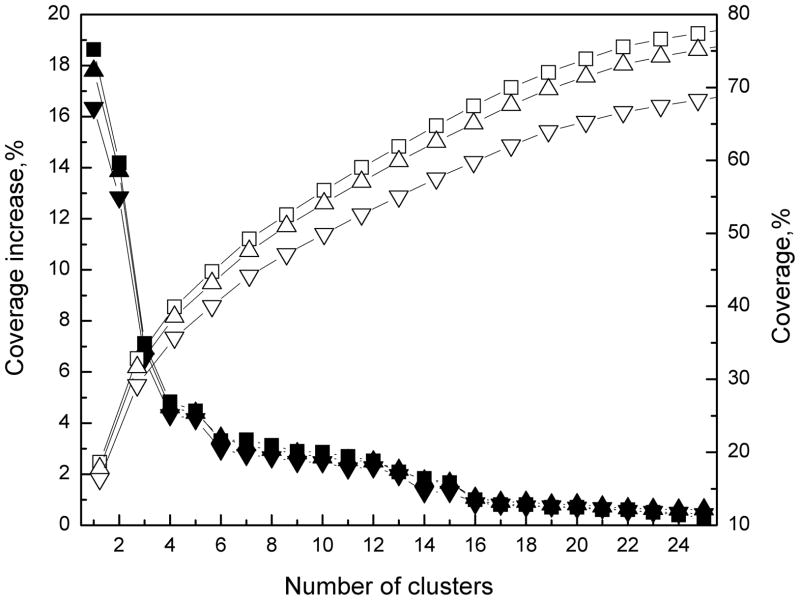
The number of the initial clusters for the long side chains was selected in such a way that the coverage gains < 0.5% if another cluster is taken into account. An example for Lys is shown in Figure 2. According to the data in the figure, the number of clusters was 21. To optimize the coverage, for Met, we used r = 45°, and for Lys, Gln and Arg, r = 50°. In the case of Glu, an ellipsoid with the radii (50°, 50°, 40°) was used instead of a sphere.
Figure 2. Cluster coverage of unbound interface Lysine conformations.
The coverage and the increase in coverage are shown as functions of the number of clusters, for different cluster radii: 50° (□/■), 45° (△/▲), and 35 ° (▽/▼).
Clustering in the Cartesian space
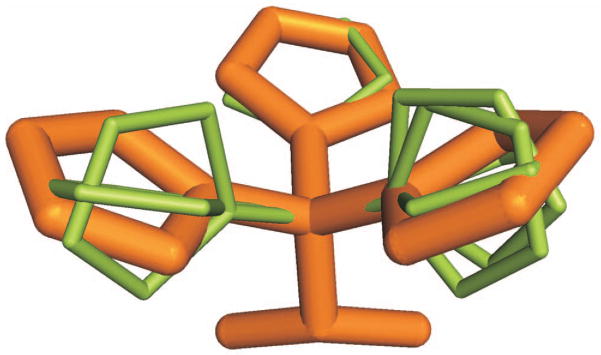
For further analysis, the rotamers dihedral angles coordinates were converted into the Cartesian coordinates. Some rotamers appeared to be very close to each other in the Cartesian space (see an example in Figure 3). Typically they differed in the last dihedral angle, which showed to be more variable.26 To remove the redundancy, the RMSD-based linkage clustering was performed on the libraries for residues with more than one dihedral angle, excluding Pro. Rotamers within a predefined radius were merged, and the rotamer with higher probability was kept as the representative. Table S4 shows the number of rotamers for different RMSD clustering radii. The distance between the rotamers was calculated as the RMSD of the side-chain atoms. To generate the non-redundant libraries (Table S3), we chose 2 Å radius often used to evaluate the accuracy of small ligand docking.32–34 After visual inspection, Leu, Lys, Met, Arg, and Gln rotamers were clustered with a slightly larger 2.3 Å radius to connect rotamers that have similar near-backbone dihedral angles and RMSD ≤ 2.3Å. The number of rotamers in the non-redundant libraries is shown in Table II. All rotamers were examined for internal clashes, with no clashes detected. A clash was defined as a distance between two non-bonding atoms < 2 Å.
Figure 3. Rotamers of Histidine.
The rotamers obtained by 2 Å RMSD clustering are in orange.
Table II.
Number of rotamers in libraries of surface residues.
| Amino acid | Redundant Library | Non-Redundant Library | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||
| Unbound | Bound | Unbound | Bound | |||||||||
|
| ||||||||||||
| FS a | NIS b | IS c | FS | NIS | IS | FS | NIS | IS | FS | NIS | IS | |
|
| ||||||||||||
| Cys | 3 | 3 | 3 | 3 | 3 | 3s | 3 | 3 | 3 | 3 | 3 | 3 |
| Asp | 6 | 6 | 6 | 6 | 6 | 6 | 3 | 3 | 3 | 3 | 3 | 3 |
| Glu | 15 | 15 | 13 | 15 | 14 | 13 | 8 | 8 | 7 | 8 | 7 | 7 |
| Phe | 4 | 4 | 3 | 4 | 4 | 4 | 3 | 3 | 3 | 3 | 3 | 3 |
| His | 9 | 9 | 9 | 9 | 9 | 8 | 3 | 3 | 3 | 3 | 3 | 3 |
| Ile | 7 | 7 | 7 | 7 | 7 | 7 | 3 | 3 | 3 | 3 | 3 | 3 |
| Lys | 21 | 21 | 23 | 20 | 20 | 22 | 5 | 5 | 5 | 5 | 5 | 5 |
| Leu | 10 | 10 | 7 | 9 | 9 | 9 | 3 | 3 | 3 | 3 | 3 | 3 |
| Met | 10 | 10 | 6 | 10 | 10 | 6 | 5 | 5 | 4 | 5 | 5 | 3 |
| Asn | 5 | 5 | 5 | 5 | 5 | 5 | 3 | 3 | 3 | 3 | 3 | 3 |
| Pro | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
| Gln | 17 | 17 | 14 | 17 | 15 | 14 | 8 | 7 | 6 | 7 | 7 | 6 |
| Arg | 26 | 25 | 26 | 25 | 26 | 21 | 9 | 11 | 12 | 9 | 13 | 10 |
| Ser | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 |
| Thr | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 |
| Val | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 |
| Trp | 7 | 7 | 7 | 7 | 7 | 7 | 7 | 7 | 7 | 7 | 7 | 7 |
| Tyr | 4 | 4 | 4 | 4 | 4 | 4 | 3 | 3 | 3 | 3 | 3 | 3 |
Full surface
Non-interface surface
Interface surface
Probability of side-chain transition upon binding
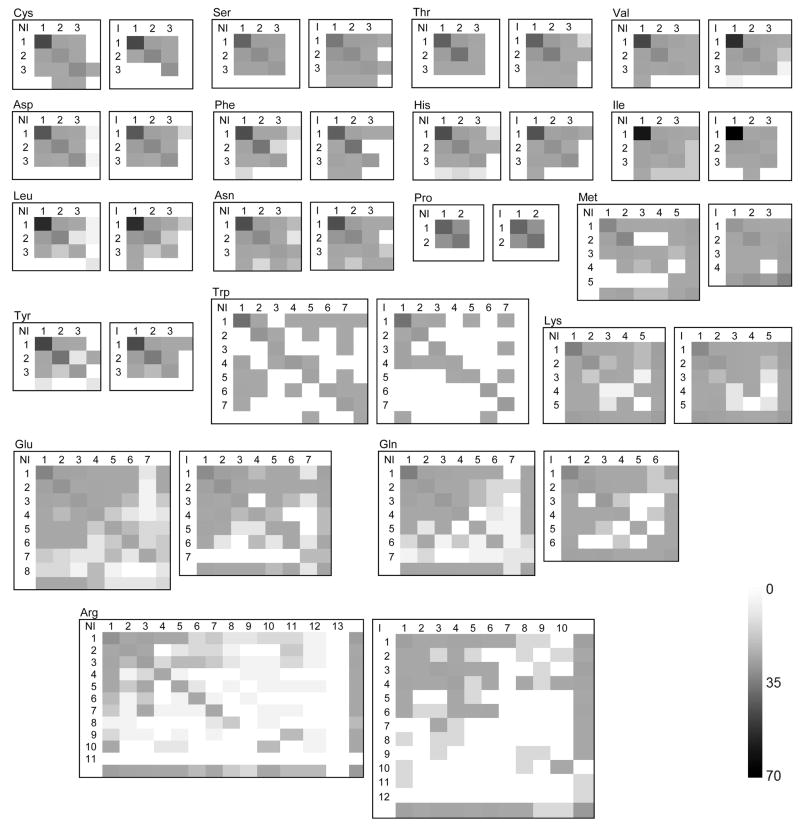
Bound-to-unbound rotamer transition maps/matrices (Tables S5, S6 and Figure 4), containing percentages and probabilities of transition between rotamers and within rotamers, were compiled for each amino acid at the interface and non-interface areas. The rows and columns of the transition maps/matrices correspond to a rotamer in the unbound (row) and bound (column) states. The non-redundant libraries of bound/unbound interface/non-interface rotamers (Table S3) were used. A transition matrix element (i,j) is the probability of the conformational change from unbound rotamer i to bound rotamer j calculated as a ratio between the number of i to j transitions and all transitions of the unbound rotamer i. The element (i,j) of a transition map is the percentage of changes between unbound rotamer i and bound rotamer j in all conformational changes of the amino acid. The rows and columns with no number in Tables S5 and S6 correspond to a set of conformations not assigned to rotamers. The corresponding matrix element is the percentage/probability of a transition, where one or both side-chains are in the non-rotameric conformation. The sum of all elements in the transition map, as well as the sum of the elements in each line in the transition matrix, is 100.
Figure 4. Unbound-to-bound conformational transition maps.
The element tij is the percentage of changes between unbound rotamer i and bound rotamer j in all conformational changes of the amino acid (Σtij = 100). The rows and columns with no number correspond to conformations not assigned to rotamers. The numerical values are in Table S5.
The rotamer stability can be evaluated by a corresponding diagonal element of the transition matrix that corresponds to the probability of a conformational change within the same rotamer. The overall stability of an amino acid is the sum of the diagonal elements of the transition map (Figure 5).
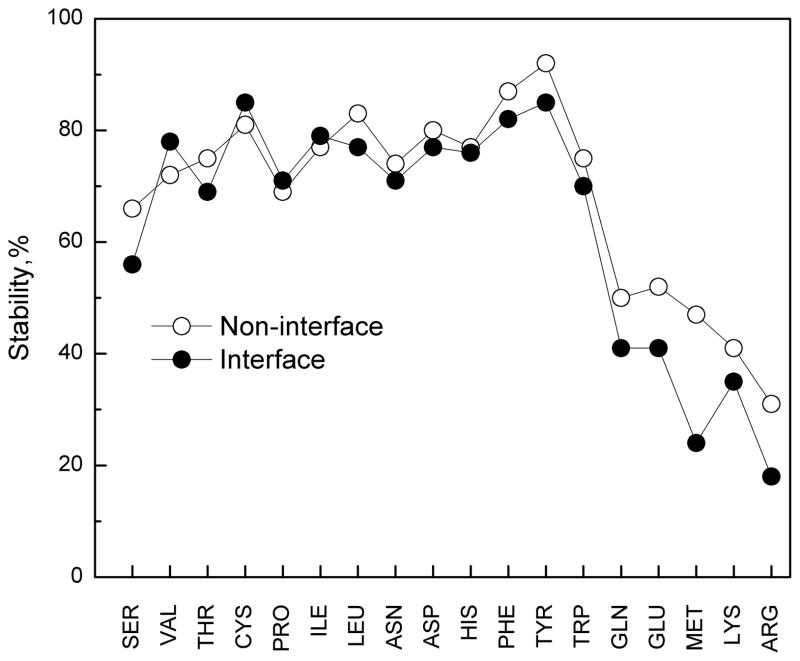
Figure 5. Stability of surface side chains.
The stability is defined as the sum of diagonal elements in transition maps.
RESULTS AND DISCUSSION
To explore the conformational preferences of bound and unbound surface side chains at the interface and non-interface areas, six rotamer libraries were generated: full unbound, interface unbound, non-interface unbound, full bound, interface bound, and non-interface bound (Tables S2 and S3).
Rotamer libraries
Table II shows the number of rotamers in the redundant and non-redundant rotamer libraries. The variation of rotamer numbers for Glu, Leu, Met, Gln, Lys, and Arg between the rotamer libraries results from disappearance of the low probability rotamers (see Tables S2 and S3). The libraries include dihedral angle values for each rotamer, the number of conformations in a cluster associated with the rotamer, and the probability of the rotamer. Comparison of the rotamer libraries is summarized in Table III, showing the RMSD in the dihedral angles space between the closest rotamers from different libraries, along with the difference in the rotamer probabilities/shares. The rotamers of small amino acids with one dihedral angle (Cys, Ser, Thr, Val) and Pro are similar in bound and unbound states in all three libraries (RMSD between closest rotamers ≤ 13°, difference between the rotamer probabilities ≤ 10%). The differences of RMSD > 50° were found between the rotamer libraries of Leu, His, Gln, Glu, Met and Arg. Variations in the last dihedral angle are often the main reason for the larger differences between the rotamers. The maximal difference between the rotamer probabilities is 14%. The rotamer coverage of long residues Met, Gln, Lys and Arg is smaller than the coverage of the shorter residues in all the libraries. Long amino acids with three and four dihedral angles have more degrees of freedom and thus more rotamers, as expected. The dihedral angles distributions of these residues are sparser, due to the larger number of non-rotameric residues.
Table III.
Comparison of non-redundant bound and unbound, interface and non-interface rotamer libraries.
| Amino acid | Bound – Unbound | Unbound | Bound | ||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Interface | Non-interface | Interface - Non-interface | Interface - Non-interface | ||||||||||||||||||||||||||
| Rotamer # | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | |
| SER | Δχa Δ,% b |
5 −1 |
4 −2 |
2 3 |
1 0 |
1 1 |
2 −1 |
−4 −7 |
1 1 |
2 4 |
2 −6 |
4 4 |
−2 1 |
||||||||||||||||
| VAL | Δχ Δ,% |
4 −1 |
1 −2 |
3 4 |
0 −1 |
2 1 |
6 1 |
4 −6 |
1 2 |
9 3 |
0 −6 |
0 4 |
0 1 |
||||||||||||||||
| THR | Δχ Δ,% |
7 3 |
2 −2 |
0 −1 |
0 2 |
2 −3 |
2 1 |
2 3 |
2 −3 |
0 −2 |
6 1 |
−2 −4 |
3 0 |
||||||||||||||||
| CYS | Δχ Δ,% |
4 0 |
2 −2 |
13 4 |
4 5 |
1 3 |
4 −7 |
4 −10 |
5 5 |
10 5 |
4 −4 |
8 9 |
1 −5 |
||||||||||||||||
| PRO | Δχ Δ,% |
3 0 |
3 0 |
2 −1 |
1 1 |
1 −4 |
2 4 |
1 −4 |
3 4 |
||||||||||||||||||||
| ILE | Δχ Δ,% |
4 −7 |
6 6 |
12 0 |
4 1 |
3 0 |
5 −1 |
1 −5 |
5 3 |
13 3 |
2 3 |
6 −3 |
5 2 |
||||||||||||||||
| LEU | Δχ Δ,% |
5 3 |
4 −2 |
66 0 |
3 −2 |
5 2 |
103 0 |
2 7 |
3 −6 |
65 0 |
3 2 |
4 −2 |
90 0 |
||||||||||||||||
| ASN | Δχ Δ,% |
13 −2 |
24 2 |
4 −3 |
3 −1 |
10 0 |
24 0 |
3 −1 |
10 2 |
5 −6 |
13 0 |
12 1 |
27 −3 |
||||||||||||||||
| ASP | Δχ Δ,% |
11 −1 |
9 3 |
10 −2 |
2 −1 |
48 1 |
8 1 |
12 0 |
10 3 |
3 −2 |
20 0 |
45 0 |
17 0 |
||||||||||||||||
| HIS | Δχ Δ,% |
166 −4 |
10 2 |
11 2 |
9 −1 |
6 1 |
7 1 |
153 −6 |
33 12 |
15 −6 |
17 −3 |
30 11 |
19 −8 |
||||||||||||||||
| PHE | Δχ Δ,% |
19 −4 |
1 6 |
10 −3 |
4 −1 |
3 3 |
8 −1 |
7 −5 |
5 1 |
15 2 |
17 −3 |
4 −2 |
14 4 |
||||||||||||||||
| TYR | Δχ Δ,% |
4 −4 |
7 3 |
1 1 |
4 5 |
3 −6 |
8 1 |
6 −5 |
9 3 |
6 2 |
8 4 |
3 −6 |
5 2 |
||||||||||||||||
| TRP | Δχ Δ,% |
12 −10 |
2 5 |
17 3 |
18 7 |
9 2 |
2 −3 |
1 −9 |
6 −1 |
2 −5 |
14 3 |
14 1 |
5 −3 |
11 3 |
3 0 |
6 −7 |
8 8 |
17 5 |
10 3 |
10 −1 |
12 −6 |
13 −5 |
3 2 |
9 1 |
34 3 |
11 −2 |
3 −3 |
13 −7 |
7 8 |
| Amino acid | Bound-Unbound | Unbound | Bound | |||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Interface | Interface-Non-interface | Interface-Non-interface | Interface-Non-interface | |||||||||||||||||||||||||||||||||||
| Rotamer # | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | |
| GLN | Δχ Δ,% |
115 0 |
12 2 |
106 −4 |
16 3 |
148 0 |
17 −2 |
6 4 |
5 1 |
116 0 |
18 −1 |
113 0 |
29 −1 |
21 0 |
104 −6 |
12 1 |
109 −4 |
21 0 |
133 0 |
35 0 |
117 −1 |
13 −2 |
16 0 |
114 1 |
21 −4 |
122 0 |
28 1 |
116 −1 |
||||||||||
| GLU | Δχ Δ,% |
18 1 |
8 4 |
32 −5 |
16 2 |
6 2 |
50 0 |
14 −2 |
49 0 |
7 −2 |
9 0 |
30 −4 |
47 0 |
71 1 |
11 1 |
11 −1 |
3 6 |
19 −4 |
25 −1 |
39 0 |
62 −1 |
9 −2 |
58 −2 |
10 0 |
17 1 |
21 −6 |
75 0 |
34 −2 |
18 1 |
|||||||||
| MET | Δχ Δ,% |
32 −10 |
135 −14 |
151 3 |
7 −5 |
154 −8 |
28 7 |
8 1 |
17 −4 |
33 −13 |
33 −11 |
35 1 |
93 3 |
26 −8 |
14 −5 |
143 5 |
||||||||||||||||||||||
| LYS | Δχ Δ,% |
4 −4 |
5 5 |
9 −2 |
5 1 |
27 2 |
1 0 |
2 1 |
6 1 |
10 2 |
11 −2 |
5 −1 |
6 2 |
11 0 |
10 0 |
15 1 |
6 3 |
6 −2 |
10 3 |
6 2 |
23 −3 |
|||||||||||||||||
| ARG | Δχ Δ,% |
13 3 |
62 3 |
63 2 |
18 −1 |
104 −2 |
13 −1 |
20 0 |
39 0 |
36 −1 |
8 3 |
9 −1 |
74 1 |
12 1 |
15 1 |
9 1 |
16 0 |
24 0 |
45 0 |
28 0 |
0 0 |
14 −11 |
126 9 |
22 7 |
178 −10 |
11 1 |
30 1 |
28 −3 |
51 0 |
35 −1 |
96 −1 |
22 −11 |
111 7 |
12 6 |
17 −8 |
21 2 |
29 −2 |
36 0 |
RMSD in the dihedral angles space ( ) between closest rotamers in the two libraries
Difference between rotamer shares in the two libraries
There is a significant difference between the non-redundant libraries of Arg and Lys. Both amino acids have long, positively charged side chains with four dihedral angles. In all six libraries, Lys has five rotamers that cover 77 – 81% of all residues (Table IV) and are very similar in conformation and probability. In contrast, Arg has 9 – 13 rotamers in different libraries that cover 67 – 77% of all residues. The conformations of the corresponding rotamers have larger variation between the libraries, especially between the interface and non-interface bound/unbound libraries (Table III). Such significant difference between Arg and Lys may be explained by the choice of the clustering RMSD, different sizes of the terminal group, and different statistics (in our protein sets, Lys is represented ~1.6 times more than Arg). Also Arg and Lys show different conservation propensities.18 Arg is highly conserved at the interface and to a lesser extent at the non-interface. At the same time, Lys is weakly conserved at the interface and highly conserved at the non-interface.
Table IV.
Coverage of rotamer libraries.
| Amino acid | Interface Unbound/Bound | Non-interface Unbound/Bound | Full Unbound/Bound | Penultimate rotamer library47 |
|---|---|---|---|---|
|
| ||||
| SER | 98/99 | 100/100 | 100/100 | 98 |
| VAL | 98/98 | 100/99 | 100/100 | 99 |
| THR | 98/98 | 100/100 | 100/100 | 99 |
| CYS | 100/98 | 100/98 | 100/97 | 99 |
| PRO | 100/100 | 100/100 | 100/100 | 93 |
| ILE | 99/100 | 98/98 | 98/98 | 99 |
| LEU | 100/99 | 100/100 | 100/100 | 93 |
| ASN | 91/95 | 96/97 | 96/97 | 94 |
| ASP | 100/100 | 100/100 | 100/100 | 96 |
| HIS | 99/98 | 99/99 | 99/99 | 94 |
| PHE | 98/99 | 100/100 | 99/99 | 98 |
| TYR | 100/99 | 99/99 | 100/99 | 98 |
| TRP | 94/98 | 97/97 | 95/97 | 94 |
| GLN | 82/83 | 91/89 | 91/91 | 88 |
| GLU | 92/90 | 96/97 | 96/97 | 91 |
| MET | 51/66 | 79/81 | 75/82 | 86 |
| LYS | 81/79 | 79/77 | 78/78 | 81 |
| ARG | 75/67 | 76/74 | 77/74 | 82 |
Overall, the small difference between bound and unbound rotamers should be expected because the majority of proteins in the current non-redundant sets of protein complexes (e.g., Dockground set and the Benchmark set35 from Weng and co-workers) undergo small unbound-to-bound conformational changes. Indeed, The Weng’s Benchmark has interface Cα RMSD < 2.2 Å for 86% of complexes, and the Dockground set used in this study has Cα RMSD < 2 Å for 71% of complexes.
Inter- and intra-rotamer transitions
Analysis of the transition maps/matrices (Figure 4 and Tables S5/S6) shows that, in general, large numbers are on the diagonal, indicating that a conformation in the unbound state prefers to stay in the same rotamer in the bound state. There are, however, some exceptions related to the rotamers with small occupancy. Large elements of the transition maps/matrices were found also for transitions to a bound rotamer adjacent to the unbound one. In such case, the bound rotamer differs slightly from the unbound one in the near-backbone dihedral angle.
The percentage of the transitions between rotamers often decreases with the decrease of the rotamer occupancy (Table S5). There are few exceptions in the case of long residues as well as Trp and Ser. The percentage of the transitions between the two most occupied rotamers is usually higher than the percentage of other transitions. Interestingly, this is true also when it involves changes in the first dihedral angle, even though such changes are rare and may cause large conformational shifts not characteristic to rotameric transitions. The probability of the transitions between the rotamers increased with the decrease of the rotamer stability (Table S6).
The stabilities of interface and non-interface residues (the trace of the transition matrix) are shown in Figure 5. The stability on the non-interface surface is almost always higher than that at the interface, reflecting the fact that the non-interface surface residues are not directly affected by the binding, and the influence of the crystal contacts is weaker than that of the interface contacts (see also Ref.26). Whether at the interface or on non-interface surface, long residues with three or four dihedral angles were always less stable than the shorter residues. The interface stability larger than the non-interface one by > 9% was determined for Ser, Gln, Glu, Met, and Arg. High conformation flexibility of long residues and Ser on protein surface was also observed in the ensembles of unbound proteins, possibly due to crystal packing.36
Comparison with other rotamer libraries
A number of unbound rotamer libraries have been published previously 25,30,37–48 (see Ref.49 for a review). There are backbone-independent, secondary-structure-dependent, and backbone-dependent rotamer libraries. In comprehensive analysis of rotamers by Dunbrack and Cohen25 the torsional space was divided into bins, and Bayesian statistics was used to obtain population estimates of sparse regions. In one of the latest backbone-independent rotamer libraries, the “Penultimate rotamer library” (PL)47 by Lovell, Richardson and co-workers, the dihedral angle space was clustered and rotamer positions were defined as the distribution mode. Generally, the rotamer libraries are constructed by either clustering the observed conformations or by dividing dihedral angle space into bins and determining the most probable conformation in each bin. Because of these inherent differences in the rotamer definitions, the comparison of these two types of libraries is not straightforward. Thus, we compared our library to the PL rotamer library of Richardson et al., which is based on a similar paradigm of rotamer definition by clustering in the dihedral angles space. The comparison with our redundant full surface unbound library is detailed in Table S7.
Generally, the two libraries are similar in rotamers and their probabilities. The similarity between the libraries indicates the robustness of our results, because significantly different protein sets and clustering methods were used to derive the rotamer libraries in these studies. There are some rotamers in our library that correspond to two PL rotamers and vice versa. For example, Asn rotamer 3 in our library corresponds to three PL rotamers (m-20°, m-80° and m120°). Gln rotamer mt-30° in PL corresponds to rotamers 1, 2, and 5 in our library. Some low probability rotamers disappear altogether (e.g., Glu rotamers 10, 13, and 15 in our library, and Met rotamers ptp, tpt, ttt, mmt in PL). Corresponding rotamers may also be different in the last dihedral angle (Tyr, Gln, and Arg rotamers 4), which corresponds to the rotation of the side chain tip, and have small RMSD. Some differences in the rotamer probabilities result from different rotamer boundaries used in the libraries. The principal difference, of course, is the datasets used to compile the libraries – unbound proteins for PL and bound vs. unbound proteins in our libraries.
CONCLUSIONS AND FUTURE DIRECTIONS
Side chains in protein-protein complexes have been analyzed in bound and unbound conformations at the interface and non-interface surface areas. Six rotamer libraries were generated: full surface, surface non-interface, and surface interface - each for bound and unbound states. The rotamers represented local peaks in multidimensional distribution of conformations in the dihedral space. The Variable Threshold clustering algorithm was applied to derive the rotamers, defined as the most probable conformations in the clusters. To generate non-redundant rotamer libraries, the rotamers were further clustered with a 2 Å radius. These libraries provide an opportunity to reduce the sampling of conformational space in docking, while maintaining 2 Å accuracy. The analysis of the rotamer libraries revealed their high similarity. The rotamer libraries were used to generate maps/matrices of unbound to bound transitions for the surface side chains. The transition maps showed that, typically, most side chains change conformation within its unbound rotamer, or shift to an adjacent bound rotamer that only slightly differs from the unbound one in the near-backbone dihedral angle. The percentage of the transitions between two most occupied rotamers is usually the highest one. The interface transition maps revealed more asymmetry than the non-interface ones because of the intermolecular interaction upon binding.
Rotamer and residue stabilities were defined based on the transition maps/matrices. The non-interface residues stability was higher than that of the interface. Long side chains with three or four dihedral angles were less stable than the shorter ones. The percentage of the transitions between rotamers often decreased with the decrease of the rotamer occupancy. At the same time, the probability of the transitions between rotamers increased with the decrease of the rotamer stability.
The analysis showed differences in conformational transitions of interface and non-interface residues, which can be utilized in docking protocols. We plan to expand our study to systematically investigate the coupling between the backbone and the side-chain conformational changes at the interface and non-interface areas. This will provide a more comprehensive characterization of binding mechanisms, and may suggest more effective ways to implement protein flexibility in docking. The biased sampling based on the transition matrices may accelerate the flexible docking search by discriminating the low probability conformational states in docking approaches. Our plans involve implementation of the rotameric preferences in the flexible docking protocol, as well as comparative analysis of such preferences in experimentally determined and modeled protein structures.
Supplementary Material
Acknowledgments
The study was supported by R01 GM074255 grant from NIH. We thank Dr. Simon C. Lovell for the code converting dihedral angles to Cartesian coordinates of the atoms.
References
- 1.Janin J, Bahadur RP, Chakrabarti P. Protein-protein interaction and quaternary structure. Quart Rev Biophys. 2008;41:133–180. doi: 10.1017/S0033583508004708. [DOI] [PubMed] [Google Scholar]
- 2.Lo Conte L, Chothia C, Janin J. The atomic structure of protein-protein recognition sites. J Mol Biol. 1999;285:2177–2198. doi: 10.1006/jmbi.1998.2439. [DOI] [PubMed] [Google Scholar]
- 3.Chakrabarti P, Janin J. Dissecting protein-protein recognition sites. Proteins. 2002;47:334–343. doi: 10.1002/prot.10085. [DOI] [PubMed] [Google Scholar]
- 4.Tsai C-J, Lin SL, Wolfson H, Nussinov R. Studies of protein-protein interfaces: A statistical analysis of the hydrophobic effect. Protein Sci. 1997;6:53–64. doi: 10.1002/pro.5560060106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bosshard HR. Molecular recognition by induced fit: How fit is the concept? News Physiol Sci. 2001;16:171–173. doi: 10.1152/physiologyonline.2001.16.4.171. [DOI] [PubMed] [Google Scholar]
- 6.Smith GR, Sternberg MJE, Bates PA. The relationship between the flexibility of proteins and their conformational states on forming protein–protein complexes with an application to protein–protein docking. J Mol Biol. 2005;347:1077–1101. doi: 10.1016/j.jmb.2005.01.058. [DOI] [PubMed] [Google Scholar]
- 7.Yogurtcu ON, Erdemli SB, Nussinov R, Turkay M, Keskin O. Restricted mobility of conserved residues in protein-protein interfaces in molecular simulations. Biophys J. 2008;94:3475–3485. doi: 10.1529/biophysj.107.114835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kimura SR, Brower RC, Vajda S, Camacho CJ. Dynamical view of the positions of key side chains in protein-protein recognition. Biophys J. 2001;80:635–642. doi: 10.1016/S0006-3495(01)76044-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rajamani D, Thiel S, Vajda S, Camacho CJ. Anchor residues in protein–protein interactions. Proc Natl Acad Sci USA. 2004;101:11287–11292. doi: 10.1073/pnas.0401942101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wlodarski T, Zagrovic B. Conformational selection and induced fit mechanism underlie specificity in noncovalent interactions with ubiquitin. Proc Natl Acad Sci USA. 2009;106:19346–19351. doi: 10.1073/pnas.0906966106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goh CS, Milburn D, Gerstein M. Conformational changes associated with protein-protein interactions. Curr Opin Struct Biol. 2004;14:104–109. doi: 10.1016/j.sbi.2004.01.005. [DOI] [PubMed] [Google Scholar]
- 12.Gerstein M, Lesk A, Chothia C. Structural mechanism for domain movements in proteins. Biochemistry. 1994;33:6739–6749. doi: 10.1021/bi00188a001. [DOI] [PubMed] [Google Scholar]
- 13.Darnell SJ, Page D, Mitchell JC. An automated decision-tree approach to predicting protein interaction hot spots. Proteins. 2007;68:813–823. doi: 10.1002/prot.21474. [DOI] [PubMed] [Google Scholar]
- 14.Tobi D, Bahar I. Structural changes involved in protein binding correlate with intrinsic motions of proteins in the unbound state. Proc Natl Acad Sci USA. 2005;102:18908–18913. doi: 10.1073/pnas.0507603102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fischer E. Einfluss der Configuration auf die Wirkung der Enzyme. Ber Dt Chem Ges. 1894;27:2985–2993. [Google Scholar]
- 16.Koshland DE. Application of a theory of enzyme specificity to protein synthesis. Proc Natl Acad Sci USA. 1958;44:98–104. doi: 10.1073/pnas.44.2.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ma B, Kumar S, Tsai C-J, Nussinov R. Folding funnels and binding mechanisms. Protein Eng. 1999;12:713–721. doi: 10.1093/protein/12.9.713. [DOI] [PubMed] [Google Scholar]
- 18.Ma B, Elkayam T, Wolfson H, Nussinov R. Protein-protein interactions: Structurally conserved residues distinguish between binding sites and exposed protein surfaces. Proc Natl Acad Sci USA. 2003;100:5772–5777. doi: 10.1073/pnas.1030237100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Betts MJ, Sternberg MJE. An analysis of conformational changes on protein-protein association: Implications for predictive docking. Protein Eng. 1999;12(4):271–283. doi: 10.1093/protein/12.4.271. [DOI] [PubMed] [Google Scholar]
- 20.Najmanovich R, Kuttner J, Sobolev V, Edelman M. Side-chain flexibility in proteins upon ligand binding. Proteins. 2000;39:261–268. doi: 10.1002/(sici)1097-0134(20000515)39:3<261::aid-prot90>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 21.Zhao S, Goodsell DS, Olson AJ. Analysis of a data set of paired uncomplexed protein structures: New metrics for side-chain flexibility and model evaluation. Proteins. 2001;43:271–279. doi: 10.1002/prot.1038. [DOI] [PubMed] [Google Scholar]
- 22.Koch K, Zollner F, Neumann S, Kummert F, Sagerer G. Comparing bound and unbound protein structures using energy calculation and rotamer statistics. In Silico Biol. 2002;2:351–368. [PubMed] [Google Scholar]
- 23.Beglov D, Hall D, Brenke R, Shapovalov MV, Dunbrack RL, Kozakov D, Vajda S. Minimal ensembles of side chain conformers for modeling protein-protein interactions. Proteins. 2011;80:591–601. doi: 10.1002/prot.23222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guharoy M, Janin J, Robert CH. Side-chain rotamer transitions at protein–protein interfaces. Proteins. 2010;78:3219–3225. doi: 10.1002/prot.22821. [DOI] [PubMed] [Google Scholar]
- 25.Dunbrack RL, Cohen FE. Bayesian statistical analysis of protein side-chain rotamer preferences. Protein Sci. 1997;6:1661–1681. doi: 10.1002/pro.5560060807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ruvinsky AM, Kirys T, Tuzikov AV, Vakser IA. Side-chain conformational changes upon protein-protein association. J Mol Biol. 2011;408:356–365. doi: 10.1016/j.jmb.2011.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gao Y, Douguet D, Tovchigrechko A, Vakser IA. DOCKGROUND system of databases for protein recognition studies: Unbound structures for docking. Proteins. 2007;69:845–851. doi: 10.1002/prot.21714. [DOI] [PubMed] [Google Scholar]
- 28.Hubbard SJ, Thornton JM NACCESS. Computer Program, Department of Biochemistry and Molecular Biology. University College London; 1993. [Google Scholar]
- 29.Word JM, Richardson D, Richardson J. Computer Program. Duke University; Dang. [Google Scholar]
- 30.Dunbrack RL, Karplus M. Backbone-dependent rotamer library for proteins: Application to side-chain prediction. J Mol Biol. 1993;230:543–574. doi: 10.1006/jmbi.1993.1170. [DOI] [PubMed] [Google Scholar]
- 31.Heyer LJ, Kruglyak S, Yooseph S. Exploring expression data: Identification and analysis of coexpressed genes. Genome Res. 1999;9:1106–1115. doi: 10.1101/gr.9.11.1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ding F, Yin S, Dokholyan NV. Rapid flexible docking using a stochastic rotamer library of ligands. J Chem Inf Model. 2010;50:1623–1632. doi: 10.1021/ci100218t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ruvinsky AM, Kozintsev AV. Novel statistical-thermodynamic methods to predict protein-ligand binding positions using probability distribution functions. Proteins. 2006;62:202–208. doi: 10.1002/prot.20673. [DOI] [PubMed] [Google Scholar]
- 34.Huey R, Morris GM, Olson AJ, Goodsell DS. A semiempirical free energy force field with charge-based desolvation. J Comput Chem. 2007;28:1145–1152. doi: 10.1002/jcc.20634. [DOI] [PubMed] [Google Scholar]
- 35.Hwang H, Pierce B, Mintseris J, Janin J, Weng Z. Protein–protein docking benchmark version 3.0. Proteins. 2008;73:705–709. doi: 10.1002/prot.22106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jacobson MP, Friesner RA, Xiang Z, Honig B. On the role of the crystal environment in determining protein side-chain conformations. J Mol Biol. 2002;320:597–608. doi: 10.1016/s0022-2836(02)00470-9. [DOI] [PubMed] [Google Scholar]
- 37.Tuffery P, Etchebest C, Hazout S, Lavery R. A new approach to the rapid determination of protein side-chain conformations. J Biomol Struc & Dynamics. 1991;8:1267–1289. doi: 10.1080/07391102.1991.10507882. [DOI] [PubMed] [Google Scholar]
- 38.Benedetti E, Morelli G, Nemethy G, Scheraga HA. Statistical and energetic analysis of side-chain conformations in oligopeptides. Int J Pept Prot Res. 1983;22:1–15. doi: 10.1111/j.1399-3011.1983.tb02062.x. [DOI] [PubMed] [Google Scholar]
- 39.Chandrasekaran R, Ramachandran GN. Studies on the conformation of amino acids. XI. Analysis of the observed side group conformation in proteins. Int J Protein Res. 1970;2:223–233. [PubMed] [Google Scholar]
- 40.Bhat TN, Sasisekharan V, Vijayan M. Analysis of side-chain conformation in proteins. Int J Pept Prot Res. 1979;13:170–184. doi: 10.1111/j.1399-3011.1979.tb01866.x. [DOI] [PubMed] [Google Scholar]
- 41.Janin J, Wodak S. Conformation of amino acid side-chains in proteins. J Mol Biol. 1978;125:357–386. doi: 10.1016/0022-2836(78)90408-4. [DOI] [PubMed] [Google Scholar]
- 42.Ponder JW, Richards FM. Tertiary templates for proteins. Use of packing criteria in the enumeration of allowed sequences for different structural classes. J Mol Biol. 1987;193:775–791. doi: 10.1016/0022-2836(87)90358-5. [DOI] [PubMed] [Google Scholar]
- 43.Mcgregor MJ, Islam SA, Sternberg MJE. Analysis of the relationship between side-chain conformation and secondary structure in globular-proteins. J Mol Biol. 1987;198:295–310. doi: 10.1016/0022-2836(87)90314-7. [DOI] [PubMed] [Google Scholar]
- 44.Schrauber H, Eisenhaber F, Argos P. Rotamers: to be or not to be? An analysis of amino acid side-chain conformations in globular proteins. J Mol Biol. 1993;230:592–612. doi: 10.1006/jmbi.1993.1172. [DOI] [PubMed] [Google Scholar]
- 45.Kono H, Doi J. A new method for side-chain conformation prediction using a Hopfield network and reproduced rotamers. J Comput Chem. 1996;17:1667–1683. [Google Scholar]
- 46.DeMaeyer M, Desmet J, Lasters I. All in one: A highly detailed rotamer library improves both accuracy and speed in the modelling of sidechains by dead-end elimination. Folding & Design. 1997;2:53–66. doi: 10.1016/s1359-0278(97)00006-0. [DOI] [PubMed] [Google Scholar]
- 47.Lovell SC, Word JM, Richardson JS, Richardson DC. The penultimate rotamer library. Proteins. 2000;40:389–408. [PubMed] [Google Scholar]
- 48.Shapovalov MS, Dunbrack RL. A smoothed backbone-dependent rotamer library for proteins derived from adaptive kernel density estimates and regressions. Structure. 2011;19:844–858. doi: 10.1016/j.str.2011.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dunbrack RL. Rotamer libraries in the 21st century. Curr Opin Struct Biol. 2002;12:431–440. doi: 10.1016/s0959-440x(02)00344-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.