Abstract
The composite human microbiome of Western populations has likely changed over the past century, brought on by new environmental triggers that often have a negative impact on human health1. Here we show that consumption of a diet high in saturated (milk derived)-fat (MF), but not polyunsaturated (safflower oil)-fat (PUFA), changes the conditions for microbial assemblage and promotes expansion of a low abundance, sulfite-reducing pathobiont, Bilophila wadsworthia2. This was associated with a pro-inflammatory TH1 immune response and increased incidence of colitis in genetically susceptible IL-10−/−, but not wild type mice. These effects are mediated by MF-promoted taurine-conjugation of hepatic bile acids, which increases the availability of organic sulfur used by sulfite-reducing microbes like B. wadsworthia. When mice were fed a low-fat (LF) diet supplemented with taurocholic, but not with glycocholic acid, for example, a bloom of B. wadsworthia and development of colitis were observed in IL10−/− mice. Together these data show that dietary fats, by promoting changes in host bile acid composition, can dramatically alter conditions for gut microbial assemblage, resulting in dysbiosis that can perturb immune homeostasis. The data provide a plausible mechanistic basis by which Western type diets high in certain saturated fats might increase the prevalence of complex immune-mediated diseases like inflammatory bowel diseases in genetically susceptible hosts.
Inflammatory bowel diseases (IBD), and other immune-related human disorders, are relatively “new” diseases in that their incidence has increased significantly over the last half century, matching developments in cultural westernization1,3,4. The rapidity of these developments are not likely caused by genetic drift, but by exposure to non-genetic factors introduced through changes in diet and lifestyle of genetically susceptible individuals, triggering aberrant host responses that lead to IBD. In this study, we examined if certain dietary fats represented in Western diets are capable of precipitating colonic inflammation through their actions on the enteric microbiota of genetically-susceptible hosts.
The effects of three different diets (Supplementary Table 1) on the enteric microbiota of specific pathogen-free (SPF) C57BL/6 mice are shown in Fig 1a and Supplementary Table 2. With the exception of the LF purified mouse diet, the high fat diets were isocaloric and differed only in the type of dietary fat used which was held constant at 37% of total calories and closely mimics Western consumption5. These fats also represent sources used in numerous processed and confectionary foods. Twenty-one-day exposure to the three study diets resulted in significant differences in the structure of the enteric microbiota as assessed by both Sanger-based and 454-based DNA sequencing of 16S rRNA libraries from cecal contents and stool. Both high-fat diets reduced the richness of the microbiota compared with LF (Supplementary Fig. 1). LF promoted Firmicutes, but also had a lower abundance of most other phyla, whereas PUFA and MF resulted in a higher abundance of Bacteroidetes and lower abundance of Firmicutes. Interestingly, these changes differed from those induced by lard-based, saturated fats6,7 (Supplementary Fig. 2). While MF and PUFA had similar effects on Bacteroidetes and Firmicutes, a significant bloom of a member of the Deltaproteobacteria, Bilophila wadsworthia, was consistently observed only with MF. B. wadsworthia is a sulfite-reducing, immunogenic microbe that is difficult to detect in healthy individuals, but emerges under pathological conditions such as appendicitis and other intestinal inflammatory disorders 8–16.
Figure 1. Saturated MF-induced colitis is associated with bloom of Bilophila wadsworthia (Bw) in IL10−/− mice.
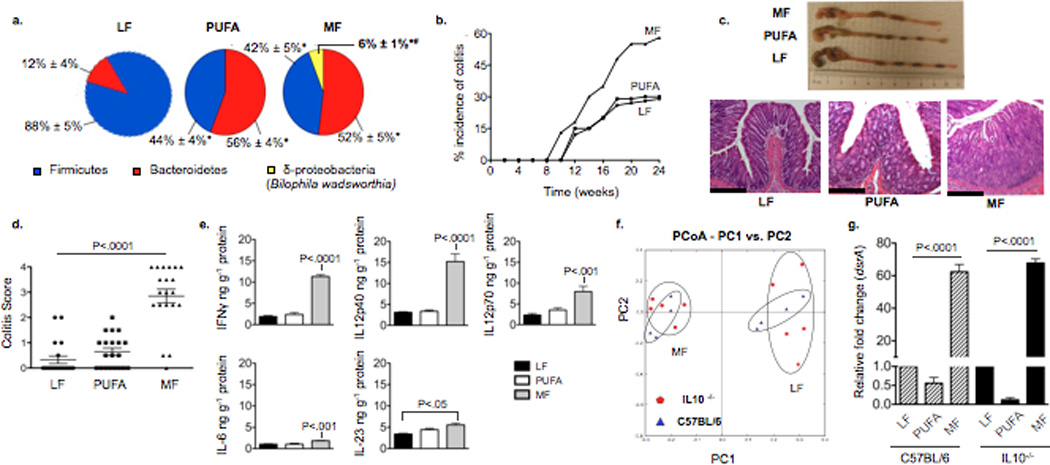
a,g Samples from SPF C57BL/6 (n=6/group), and b–g, SPF IL10−/− mice fed MF, PUFA or LF for 24 weeks (n=20/group). a, Phyla representation shown for LF, PUFA, and MF with means ± S.E.M. *P<.05 compared to LF, #P<.05 compared to PUFA and LF. b, Gross incidence of colitis. c, Representative colon lengths (top) and H&E staining of distal colon (bottom). Scale bars, 400µm. d, Blinded histological colitis scores25. e, Distal colonic mucosal cytokines determined by ELISA. f, PCoA plot of the UNIFRAC metric matrix. g, Q-PCR of cecal content dsrA (normalized to LF diet).
MF did not affect wild-type mice, but increased the onset and incidence of colitis in IL-10−/− mice, driving it from a spontaneous rate of 25–30% (on LF) to over 60% in a 6-month period (Fig. 1b). In contrast, the incidence of colitis in IL-10−/− mice fed PUFA was no different than those fed LF. The colitis seen in mice fed MF was also more severe and extensive (Fig. 1c). These changes paralleled differences in histological colitis scores (Fig. 1d). Inflammatory mucosal cytokine levels from the distal colon were significantly elevated compared to LF and in most cases, PUFA (Fig. 1e). LF, PUFA, and MF elicited effects on the total enteric microbiota of IL-10−/− mice that were similar to those observed in wild-type mice (shown for LF and MF, Fig. 1f). Similarly, B. wadsworthia, as detected by qPCR of the dissimilatory sulfite reductase A gene unique to sulfite-reducing bacteria of which B. wadsworthia is the most prominent in our model, was found at equal relative abundance on MF independent of genotype (Fig. 1g). The bloom of B. wadsworthia induced by MF was also observed in DSS-treated SPF C57BL/6 mice where the onset and severity of colitis were more severe than that seen in LF and PUFA-fed mice (Supplementary Figure 3). Altogether these observations suggest that the bloom of sulfite-reducing Deltaproteobacteria, particularly B. wadsworthia, is associated with colitis in hosts that are genetically susceptible or have compromised mucosal barrier function.
To explore whether MF was necessary for B.wadsworthia’s survival and proliferation, we monoassociated germ-free (GF) IL-10−/− mice with B. wadsworthia (ATCC 49260) that were consuming either LF, PUFA, or MF. Five-weeks post-gavage, colonization of the colon could only be established in mice fed MF (Supplementary Figure 4, shown for LF and MF), whereas on LF, B. wadsworthia was undetectable. B. wadsworthia identity was confirmed by PCR of the 16S rRNA encoding gene from cecal-derived DNA using universal primers, followed by direct sequencing of the PCR product, and cultivation of B. wadsworthia from cecal contents (Fig. 2a). Additionally, qPCR analysis of luminal vs. mucosal-associated B. wadsworthia revealed a nearly 45-fold increase in the latter (Supplementary Figure 5). MF, in contrast to LF or PUFA, increased the incidence of colitis (Fig. 2b), although to a lesser extent than that seen in SPF IL-10−/− mice (Supplementary Figure 6a). Furthermore, increased levels of IFNγ, IL12p40 and IL12p70, as well as low or undetectable levels of IL-6, IL-17, and IL-23 in the colonic mucosa of these mice, were consistent with the induction of a distinct TH1 immune response (Supplementary Figure 6b). This was further confirmed by increased CD4+ IFNγ+ populations in the mesenteric lymph nodes (MLN) of mice colonized on MF (Fig. 2c). These changes were not observed in mice consuming MF in the absence of B. wadsworthia, indicating that the diet itself is not immunogenic. Moreover, when Lactobacillus murinus, which is also promoted by MF, was monoassociated in GF IL-10−/− mice, no evidence of colitis or immune activation was seen (data not shown). The specificity of the TH1 response induced by B. wadsworthia was further elucidated by both ex vivo and in vitro challenges of immune cells using pure bacterial lysates from B. wadsworthia, L. murinus and Alistipes, the latter two representing bacteria that were promoted by MF (albeit to a lesser degree, Supplementary Table 2). Only lysate from B. wadsworthia elicited an IFNγ response in MLNs harvested from B. wadsworthia-monoassociated GF IL-10−/− mice (Fig. 2d). The response in IL-10−/− MLNs was much greater than that in C57BL/6 MLNs, most likely due to the absence of IL-10 modulating signals. An in vitro T cell differentiation assay was then performed in which MLN and splenic dendritic cells were isolated from SPF C57BL/6 mice and stimulated with pure lysates from B. wadsworthia, L. murinus, or Alistipes (+ retinoic acid and TGFβ for splenic DCs). Supernatants from these DC’s revealed significantly elevated levels of IL12p40 only in the B. wadsworthia stimulated DCs (Fig. 2e, left panel). When purified T cells were then incubated for three days with DC supernatants from each respective treatment and analyzed for IFNγ and IL-17. MLN and splenic T cells incubated with supernatants from DCs exposed to B. wadsworthia produced nearly two-fold higher levels of IFNγ compared to T cells incubated with supernatants of DCs exposed to Alistipes or L. murinus, (Fig. 2e, right panel), In accordance with in vivo data, T cells did not produce IL-17. Therefore, B. wadsworthia appears to activate DCs in a way that selectively induces a TH1-mediated colitis. Of note, the possibility that other B. wadsworthia byproducts (e.g. H2S, secondary bile acids) also stimulate DCs cannot be excluded.
Figure 2. Bw monoassociation in germ-free (GF) IL10−/− mice can only be established with consumption of MF diet, resulting in a TH1 immune response and development of colitis.
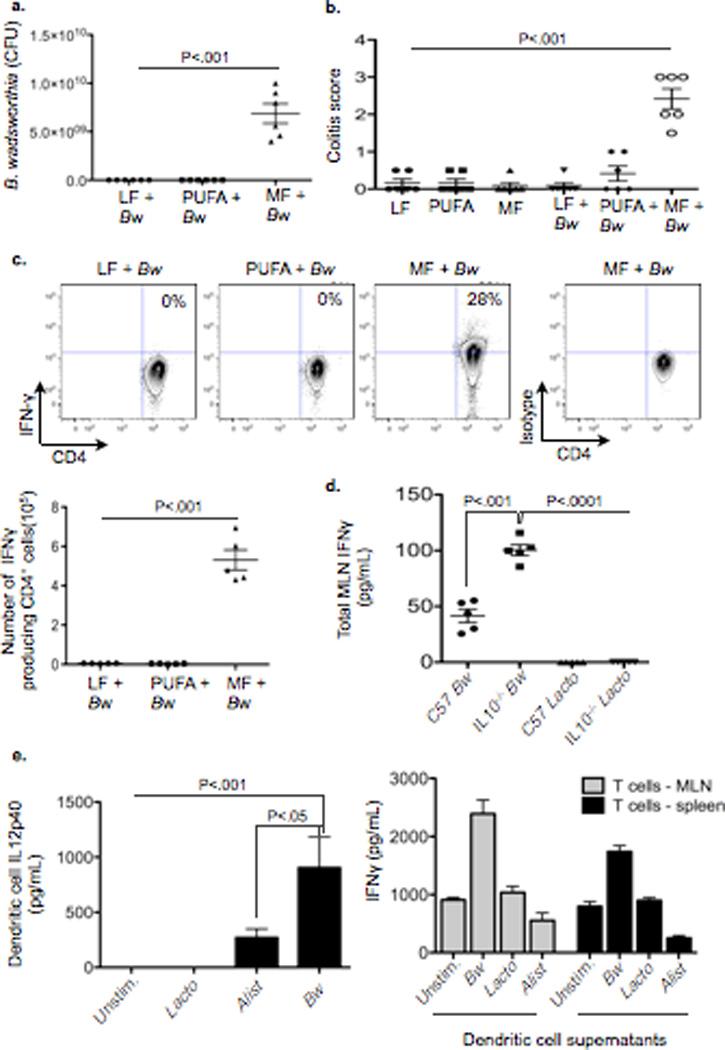
a–c, Samples from GF IL10−/− mice ± monoassociation with 108 CFU Bw maintained on LF, PUFA, or MF for 5 weeks (n=5/group). a, CFU counts of cultured cecal-derived Bw. b, Blinded histological colitis scores. c, IFNγ production by CD4+ T cells in mesenteric lymph nodes (MLN). d, IFNγ production by MLNs from GF C57BL/6 and IL10−/− mice colonized with either Bw or L. murinus (Lacto) and restimulated ex vivo with pure culture lysate from the respective bacterium. e, In vitro CD4+ T cell differentiation assay. (Left) IL12p40 produced by dendritic cells (DCs) challenged with pure lysates from Bw, Lacto, or Alistipes (Alist). Data represents pooled values from MLN DCs and splenic DCs in the presence of RA/TGFβ. (Right) IFNγ production by CD4+ T cells stimulated with supernatants from the bacteria-challenged DC’s. Data shown represents 1 out of 2 assays, performed in triplicate.
B. wadsworthia flourishes in the presence of taurine-conjugated (TC) bile acid (a property from which it got its name), a rich source of organic sulfur, which is used as the terminal electron acceptor of the electron transport chain resulting in the formation of H2S as a byproduct17. Because of their hydrophobicity, milk fats will promote increased hepatic taurine conjugation of bile acids which are more efficient for micelle formation and fat emulsification18–20, This was confirmed by mass spectrometry measurements of gall bladder aspirates from mice fed LF, PUFA, and MF (Fig 3a). When 107 CFU B. wadsworthia in taurine-free liquid growth media supplemented with 20ul of gall bladder aspirates obtained from SPF C57BL/6 mice fed the three test diets (n=5 pooled). B. wadsworthia growth was selectively and robustly stimulated only by bile from MF-fed mice (Fig. 3b). To determine whether the dietary effect was in fact mediated by TC, SPF IL-10−/− mice were fed LF and gavaged with either TC or GC daily for two weeks. This resulted in a bloom of B. wadsworthia with TC (Fig. 3c), nearly identical to what was observed with consumption of MF. In contrast, GC and PBS had little effect and B. wadsworthia remained undetectable (Fig. 3d). The bloom of B. wadsworthia observed in TC-gavaged mice was associated with increased incidence and severity of colitis (Fig. 3e and 3f). TH1 cytokines were increased in both the mucosa (Supplementary Figure 7) and MLN (Fig. 3g). In further support of the role of bile in MF diet-induced pathogenesis, monoassociation with B. wadsworthia can be established in GF IL-10−/− mice when accompanied by TC administration, but not by GC or PBS (Fig. 4a), as demonstrated by the re-isolation of B. wadsworthia from the cecal contents of TC-fed mice (note black color change indicates H2S production, left panel), and confirmed by colony counts (right panel). These mice developed colitis (Fig. 4b, Supplementary Figure 8a) and again exhibited elevated TH1 mucosal responses (Fig. 4c and Supplementary Figure 8b). Altogether these findings indicate that the bloom of B. wadsworthia promoted by these dietary factors is selectively associated with TH1 immunity.
Figure 3. Induction of taurocholic bile acid (TC) following consumption of MF promotes bloom of Bw both in vitro and in SPF IL10−/− mice resulting in colitis.
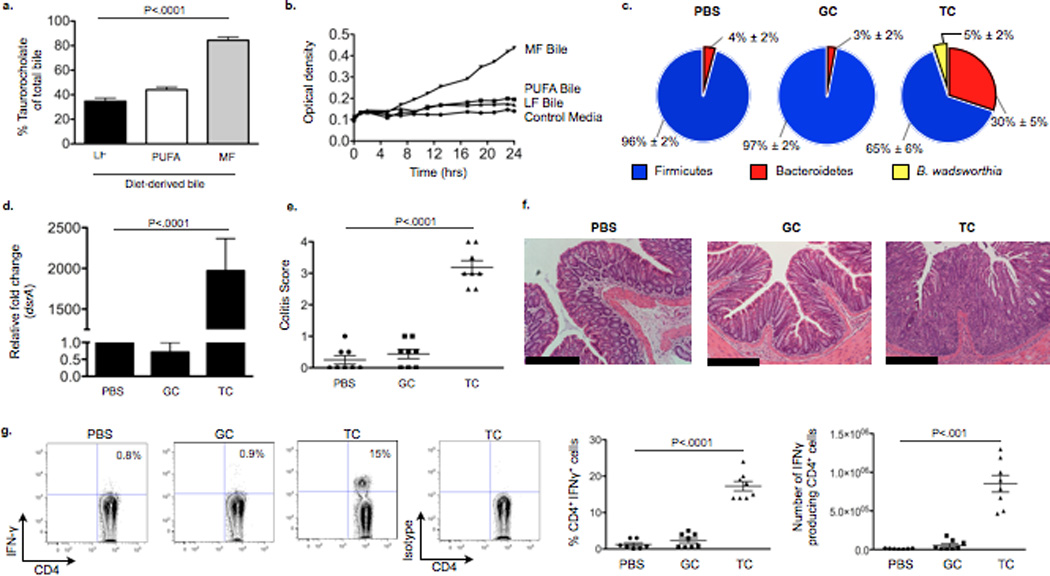
a, TC content of gall bladder aspirates from IL10−/− mice consuming LF, PUFA or MF for 5 weeks. b, Growth curve of Bw in media containing gall bladder aspirates. c–g, Samples from SPF IL10−/− mice gavaged with PBS, TC, or glycocholic acid (GC) daily for 21 days while maintained on LF diet (n=8/group). c, Phyla representation with bloom of Bw in the TC group with means ± S.E.M. *P<.05 compared to PBS and GC. d, Relative abundance of dsrA in cecal contents (by qPCR and normalized to LF diet). e, Blinded histological colitis scores, and f, H&E staining of distal colon. Scale bars, 400µm g, IFNγ production in MLNs.
Figure 4. Monoassociation with Bw in GF IL10−/− is successful only if accompanied by TC gavage.
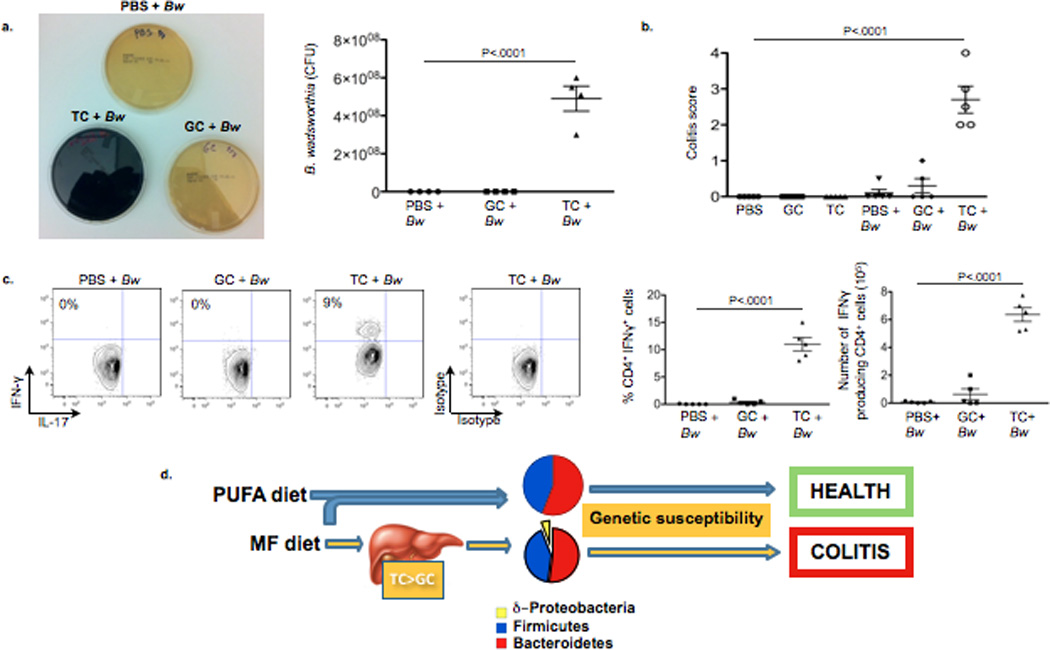
a–c, Samples from GF IL10−/− mice fed LF ± monoassociation with Bw followed by daily gavage with PBS, GC, or TC for 21 days (n=5/group). a, Robust Bw growth when re-isolated from cecal content of TC-fed GF mice (black film in TC plate indicates H2S production), and CFU counts of cecal-derived Bw. b, Blinded histological colitis scores. c, IFNγ production in MLN CD4+ T cells determined by intracellular staining. d, Proposed experimental model.
We find the dependence of B. wadsworthia on diet-induced taurocholic acid intriguing and possibly representative of how certain gut microbes utilize bile to their advantage. Bile formation is unique to vertebrates, providing the host with the ability to digest and utilize a far greater variety of dietary substrates. Bile also has potent anti-microbial properties that can contribute to the selection or exclusion of many potential gut microbiota. However, several intestinal pathogens, including protozoa such as Giardia, Microsporidia and Cryptosporidia, and bacteria such as B. wadsworthia, H. hepaticus, and L. monocytogenes, are not only bile-resistant, but highly favored in the presence of bile21, 22, possibly through suppression of symbiotic, commensal microorganisms, allowing pathobionts and pathogens an opportunity to establish a niche in the intestine. Once established, the byproducts of these bacteria, whether H2S or secondary bile acids, can serve as gut mucosal “barrier-breakers” allowing for increased immune cell infiltration and thus acts synergistically with the bacterial antigen specific immune response to induce tissue damage. In genetically susceptible hosts, this development has the capacity to tip a compensated state of immune balance in favor of chronic disease.
Methods
Mice
All mice were bred in-house and housed in our SPF and GF animal facilities. GF mice were maintained on 18% (by weight) 37% (kcal) milk fat, lard or PUFA test diets formulated by Harlan-Teklad and irradiated and tested for sterility both before and after use. Low-fat purified diet was based on AIN-93M. All bacterial gavages (Sigma-Aldrich) were performed using 108 CFU, and taurocholate and glycocholate using 1g/kg dissolved in 100ug PBS. Large particles of glycocholate were first filtered out before gavaging. All experiments were performed in accordance with the Institutional Biosafety Committee and the Institutional Care and Use Committee.
DSS Colitis
SPF C57BL/6 mice were fed LF, PUFA, or MF for 1 week (n=5/group). On day 7, 1.5% DSS was added to drinking water. On day 12, mice were changed back to plain tap water, and on day 15 mice were terminated. Weight loss and stool consistency was checked daily.
Clone library sequencing
Raw sequence data was processed using the Ribosomal Database Project’s Pipeline Tool (http://rdp.cme.msu.edu/) that includes base-calling, quality trimming and alignment as part of the workflow. Additionally, chimeric sequences were screened and removed using Bellerophon. The RDP Classifier was used to assign the 16S rRNA sequences to a hierarchical taxonomy. The program mothur was used to group sequences into operational taxonomical units (OTUs) using the furthest neighbor algorithm and a 97% sequence similarity criterion. For principal coordinates analysis (PCoA), all 16S rRNA gene sequences were imported into the ARB software package and aligned into a phylogenetic tree by neighbor joining which was used to perform clustering analysis using online UniFrac without abundance weighting. Genbank accession # JQ890637-JQ894320.
16S rRNA-based amplicon library preparation and data analysis
PCR primers used were specific for the V3-V4 region of the 16S rRNA encoding gene (Escherichia coli positions 338–802; 338F: 5’-ACTCCTACGGGAGGCAGC-3’; and equimolar amounts of 802R-A 5'-TACCRGGGTHTCTAATCC-3’, 802R-B 5'-TACCAGAGTATCTAATTC-3’, 802R-C 5'-CTACDSRGGTMTCTAATC-3’, 802R-D 5'-TACNVGGGTATCTAATCC-3’. and contained 454-specific adapter sequences as well as an 8-bp barcode. This barcode-based primer approach allowed sequencing of multiple samples in a single 454 sequencing run without the need for physical partitioning. Sequencing was performed at the High-Throughput Genome Analysis Core (HGAC; part of the Institute for Genomics & Systems Biology [IGSB]) at Argonne National Laboratory. Sequences were then trimmed and classified with the QIIME toolkit. Using the QIIME wrappers, OTUs were picked at 97% sequence identity using cdhit and a representative sequence was then chosen for each OTU by selecting the most abundant sequence in that OTU. These representative sequences were aligned using PyNAST and taxonomy was assigned to them using the RDP Classifier. The PyNAST-aligned sequences were also used to build a phylogenetic tree with FastTree and unweighted UniFrac distances then computed between all samples for additional ecological analyses, including principal coordinates analysis (PCoA).
Data are available to the public via the MG-RAST system (http://metagenomics.anl.gov/) including instant availability of the sequence data, bioinformatic analyses and tools, plus the support for export to QIIME.
Sulfite-reducing bacteria quantification
SRB were quantified using specific primers developed by Marius Vital and James Tiedje (MSU) for the dsrA gene. F: 5'-CCA ACATGCACGGYTCCA-3”, R: 5'-CGTCGA ACTTGA ACTTGA ACTTGT AGG-3’.
Bile composition analysis
Bile acid conjugates in pooled gall bladder aspirates were analyzed by HPLC as described by Rossi et al26. Conjugated bile acids were quantified in the column effluent by monitoring the absorbance at 205 nm (for the amide bond). Peaks were identified using the relative retention time of known standards. Next, electrospray mass spectrometry was performed on the pooled gall bladder aspirates using a Perkin-Elmer Sciex API-III instrument modified with a nanoelectrospray source. The instrument was operated in the negative mode with Q1 IS voltage set to 600V. The IN and ORI voltages were set to 110V and 90V respectively. Chemical identity of peaks was confirmed by the fragmentation pattern of selected ions (Q3 mode) using argon collision gas. The presence of conjugated bile salts was confirmed by selection for m/z 74 (glycine), m/z 97 (sulfate), m/z 124 (taurine).
T cell and Dendritic Cell Purification
For CD4+ T cell isolation, spleens and mesenteric lymph nodes were mechanically disrupted through a 70-µm cell strainer. CD4+ cells were isolated first by CD25 negative selection using anti-CD25 APC with anti-APC microbeads on automacs. This was followed by positive immunoselection using CD4-(L3T4) microbeads (Miltenyi Biotec).
For dendritic-cell isolation, MLN and spleen were digested with 400 units ml−1 collagenase type IV (Sigma-Aldrich). Cells were filtered, resuspended in 22.5% Optiprep (Sigma-Aldrich), overlaid with Hank’s Buffered Saline (HBS) and centrifuged at 670g for 30 min. Dendritic cells were then enriched from the interface by positive immunomagnetic selection using anti-CD11c-coated beads according to the manufacturer’s recommendations (Miltenyi Biotec). Purification yielded up to 90% CD11c+ cells.
In vitro T cell-differentiation assay
Dendritic cells from MLN and spleen + RA/TGFβ were incubated 24 hour with 25ug/mL lysate from either B. wadsworthia, Alistipes, or L. murinus. Supernatants were analyzed for IL12p40 by ELISA and diluted 25%, then applied to purified CD4+ T cells stimulated with plate bound anti-CD3 (1 µg ml−1) and anti-CD28 (2 µg ml−1) for three days. The supernatants from these cultures were then analyzed for IFNγ and IL-17.
Antibodies and flow cytometry
The following conjugated antibodies were purchased from eBioscience (San Diego): CD4 (GK1.5), CD11c (N418), IFN-γ (XMG1.2), IL-17 (eBio17B7), and isotype controls. Cells were permeabilized with the CytoFix/CytoPerm kit (BD Biosciences) for intra-cytoplasmic detection of IFN-γ and IL-17 cytokines. Flow cytometry analysis was performed with a FACsCanto (BD Biosciences).
Supplementary Material
Acknowledgements
This work was supported by the National Center for Research Resources and the NIDDK, NIGMS, and NCCAM of the National Institutes of Health through Grant Number DK42086 (EBC), DK47722 (EBC), UH3DK083993 (EBC), F31AT006073 (SD). Also, the Gastrointestinal Research Foundation, Crohns and Colitis Foundation of America (YW), the Peter and Carol Goldman Family Research Fund, and the Harry and Leona Helmsley Trust Foundation (SHARE).
We are also indebted to Sidney Finegold (UCLA) for his suggestions on successful culture of B. wadsworthia and Alistipes, James Tiedje and Marius Vital (MSU) for dsrA primer sequences, Edmond Huang, Betty Theriault, and Jennifer Stencel for assistance with experiments (UC), and Romain Bouziat (UC) for T cell purification.
Footnotes
Supplementary information is linked to the online version at www.nature.com/nature.
Contributions S.D. and E.B.C. were involved in all aspects of this study, especially in the development of the hypothesis, experimental plan, and data analysis. Y.W., M.W.M, V.L., H.F., and A.N. helped perform the experiments. D.A.A. and B.J. provided critical feedback and expertise and assisted in the analysis of data.
Competing financial interests
The authors declare no competing financial interests.
Contributor Information
Suzanne Devkota, Department of Medicine, Section of Gastroenterology, The University of Chicago, Knapp Center for Biomedical Discovery, 900 E. 57th St., Chicago, IL 60637.
Yunwei Wang, Department of Medicine, Section of Gastroenterology, The University of Chicago, Knapp Center for Biomedical Discovery, 900 E. 57th St., Chicago, IL 60637.
Mark Musch, Department of Medicine, Section of Gastroenterology, The University of Chicago, Knapp Center for Biomedical Discovery, 900 E. 57th St., Chicago, IL 60637.
Vanessa Leone, Department of Medicine, Section of Gastroenterology, The University of Chicago, Knapp Center for Biomedical Discovery, 900 E. 57th St., Chicago, IL 60637.
Hannah Fehlner-Peach, Department of Medicine, Section of Gastroenterology, The University of Chicago, Knapp Center for Biomedical Discovery, 900 E. 57th St., Chicago, IL 60637.
Anuradha Nadimpalli, Department of Medicine, Section of Gastroenterology, The University of Chicago, Knapp Center for Biomedical Discovery, 900 E. 57th St., Chicago, IL 60637.
Dionysios A. Antonopoulos, Institute for Genomics and Systems Biology, Argonne National Laboratory, 9700 S. Cass Ave., Argonne, IL 60439
Bana Jabri, Department of Medicine, Section of Gastroenterology, The University of Chicago, Knapp Center for Biomedical Discovery, 900 E. 57th St., Chicago, IL 60637.
Eugene B. Chang, Department of Medicine, Section of Gastroenterology, The University of Chicago, Knapp Center for Biomedical Discovery, 900 E. 57th St., Chicago, IL 60637
References
- 1.Walter J, Ley R. The human gut microbiome: Ecology and recent evolutionary changes. Annu Rev Microbiol. 2010;65:411–429. doi: 10.1146/annurev-micro-090110-102830. [DOI] [PubMed] [Google Scholar]
- 2.Baron EJ. Bilophila wadsworthia: a unique Gram-negative anaerobic rod. Anaerobe. 1997;3:83–86. doi: 10.1006/anae.1997.0075. [DOI] [PubMed] [Google Scholar]
- 3.Molodecky NA, Soon IS, Rabi DM, Ghali WA, Ferris M, et al. Increasing incidence and prevalence of the inflammatory bowel diseases with time, based on systematic review. Gastro. 2012;142:46–54. doi: 10.1053/j.gastro.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 4.Maslowski KM, Mackay CR. Diet, gut microbiota and immune responses. Nat Immunol. 2011;12:5–9. doi: 10.1038/ni0111-5. [DOI] [PubMed] [Google Scholar]
- 5.NHANES. Trends in intake of energy and macronutrients- Unites States 1971–2000. 2004 http://www.cdc.gov/mmwr/preview/mmwrhtml/mm5304a3.htm. [PubMed]
- 6.Turnbaugh P, Bäckhed F, Fulton L, Gordon JI. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Mic. 2008;3:213–223. doi: 10.1016/j.chom.2008.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hildebrandt MA, Hoffman C, Sherrill-Mix SA, Keilbaugh SA, Hamady M, et al. High-fat diet determines the composition of the murine gut microbiome independently of obesity. Gastro. 2009;137:1716–1724. doi: 10.1053/j.gastro.2009.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baron EJ, Summanen P, Downes J, Roberts MC, Wexler H, Finegold SM. Bilophila wadsworthia, a unique gram-negative anaerobic rod recovered from appendicitis specimens and human faeces. J Gen Microbiol. 1989;135:3405–3411. doi: 10.1099/00221287-135-12-3405. [DOI] [PubMed] [Google Scholar]
- 9.Attene-Ramos MS, Wagner ED, Plewa MJ, Gaskins HR. Evidence that hydrogen sulfide is a genotoxic agent. Mol Cancer Res. 2006;4:9–14. doi: 10.1158/1541-7786.MCR-05-0126. [DOI] [PubMed] [Google Scholar]
- 10.Loubinoux J, Bronowicji J-P, Peireira IAC, Mougenet JL, Le Faou AE. Sulphate reducing bacteria in human faeces and their association with inflammatory diseases. FEMS Microbiol Ecol. 2002;40:107–112. doi: 10.1111/j.1574-6941.2002.tb00942.x. [DOI] [PubMed] [Google Scholar]
- 11.Rowan FE, Docherty NG, Coffey JC, O’Connell PR. Sulphate-reducing bacteria and hydrogen sulphide in the etiology of ulcerative colitis. Br J Surg. 2009;2:151–158. doi: 10.1002/bjs.6454. [DOI] [PubMed] [Google Scholar]
- 12.Beech IB, Zinkevich V. Screening of sulfate-reducing bacteria in colonoscopy samples from healthy and colitic human gut mucosa. FEMS Micro Ecol. 2000;2:147–155. doi: 10.1111/j.1574-6941.2000.tb00764.x. [DOI] [PubMed] [Google Scholar]
- 13.Gibson GR, Cummings JH, Macfarlane GT. Growth and activities of sulphate-reducing bacteria in gut contents of healthy subjects and patients with ulcerative colitis. FEMS Microbiol Ecol. 1991;86:103–112. [Google Scholar]
- 14.Scanlan PD, Shanahan F, Marchesi J. Culture-independent analysis of desulfovibrios in the human distal colon of healthy, colorectal cancer and polypectomized individuals. FEMS Microbiol Ecol. 2009;2:213–221. doi: 10.1111/j.1574-6941.2009.00709.x. [DOI] [PubMed] [Google Scholar]
- 15.Arzese A, Mercuri F, Trevisan R, Menozzi G, Botta G. Recovery ofBilophila wadsworthia from clinical specimens in Italy. Anaerobe. 1997;3:219–224. doi: 10.1006/anae.1997.0076. [DOI] [PubMed] [Google Scholar]
- 16.Baron EJ, Curren M, Henderson G, Jousimies-Somer H, Lee K, et al. Bilophila wadsworthia isolates from clinical specimens. J Clin Microbiol. 1992;30:1882–1887. doi: 10.1128/jcm.30.7.1882-1884.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laue H, Denger K, Cook AM. Taurine reduction in anaerobic respiration of Bilophila wadsworthia RZATAU. Appl Environ Microb. 1997;63:2016–2021. doi: 10.1128/aem.63.5.2016-2021.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lindstedt S, Avigan J, Goodman DS, Sjovall J, Steinberg D. The effects of dietary fat on the turnover of cholic acid and on the composition of the biliary bile acids in man. J Clin Invest. 1965;44:1754–1765. doi: 10.1172/JCI105283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rueda A, Manas M, Valverde A, Fernandez JI, Naranjo JA, Martinez-Victoria E. Conjugated bile acids and intestinal flora during the preruminant stage in goat: Influence of a lamb milk replacer. Arch Physiol Biochem. 1996;104:246–251. doi: 10.1076/apab.104.2.246.12884. [DOI] [PubMed] [Google Scholar]
- 20.Graham TO, Van Thiel DH, Little JM, Lester R. Synthesis of taurocholate by rat fetal liver in organ culture: Effects of cortisol in vitro. Am J Physiol. 1979;237:E177–E184. doi: 10.1152/ajpendo.1979.237.2.E177. [DOI] [PubMed] [Google Scholar]
- 21.Ananieva O, Nilsson I, Vorobjova T, Uibo R, Wadström T. Immune responses to bile-tolerant Helicobacter species in patients with chronic liver diseases, a randomized population group, and healthy blood donors. Clin Vacc Immunol. 2002;9:1160–1164. doi: 10.1128/CDLI.9.6.1160-1164.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dussurget O, Cabanes D, Dehoux P, Lecuit M. Listeria monocytogenes bile salt hydrolase is a PrfA-regulated virulence factor involved in the intestinal and hepatic phases of listeriosis. Mol Microbiol. 2002;4:1098–1106. doi: 10.1046/j.1365-2958.2002.03080.x. [DOI] [PubMed] [Google Scholar]
- 23.Wang Y, Devkota S, Musch MW, Jabri B, Nagler, et al. Regional mucosa-associated microbiota determine physiological expression of TLR2 and TLR4 in murine colon. PLoS One. 2010;5(10):e13607. doi: 10.1371/journal.pone.0013607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Y, Hoenig JD, Malin K, Qamar S, Petrof EO, et al. 16S rRNA gene-based analysis of fecal microbiota from preterm infants with and without necrotizing enterocolitis. ISME. 2009;3:944–954. doi: 10.1038/ismej.2009.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Berg DJ, Davidson N, Kuhn R, Muller W, Menon S, et al. Enterocolitis and colon cancer in interleukin-10-deficient mice are associated with aberrant cytokine production and CD4+ TH1- like responses. 1996;4:1010–1020. doi: 10.1172/JCI118861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rossi SS, Converse JL, Hofmann AF. High pressure liquid chromatographic analysis of conjugated bile acids in human bile: simultaneous resolution of sulfated and unsulfated lithocholyl amidates and the common conjugates bile acids. J Lipid Res. 1987;28:589–595. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


