Abstract
Tail-interacting protein of 47 kDa (TIP47) has two putative functions: lipid biogenesis and mannose 6-phosphate receptor recycling. Progress in understanding the molecular details of these two functions has been hampered by the lack of structural data on TIP47, with a crystal structure of the C-terminal domain of the mouse homologue constituting the only structural data in the literature so far. Our studies have first provided a strategy to obtain pure monodisperse preparations of the full-length TIP47/perilipin-3 protein, as well as a series of N-terminal truncation mutants with no exogenous sequences. These constructs have then enabled us to obtain the first structural characterization of the full-length protein in solution. Our work demonstrates that the N-terminal region of TIP47/perilipin-3, in contrast to the largely helical C-terminal region, is predominantly β-structure with turns and bends. Moreover, we show that full-length TIP47/perilipin-3 adopts an extended conformation in solution, with considerable spatial separation of the N- and C-termini that would likely translate into a separation of functional domains.
Keywords: circular dichroism, multiangle laser light scattering, oligomerization state, small-angle X-ray scattering, tail-interacting protein of 47 kDaa
INTRODUCTION
Perilipin, adipophilin, and the tail-interacting protein of 47 kDa (TIP47), recently redesignated as perilipin-1, perilipin-2, and perilipin-3,1 respectively, are the founding members of the PAT (perilipin/ADRP/TIP47) family of proteins, a family that functions predominantly in lipid droplet biogenesis.2 However, the precise intracellular role of TIP47/perilipin-3 is debated. TIP47/perilipin-3 has been implicated in cellular processes as diverse as mannose 6-phosphate receptor (M6PR) recycling, rhodopsin photobleaching, and viral infection, in addition to lipid droplet biogenesis.3-8 TIP47/perilipin-3 was first isolated from human placenta4 but has subsequently been found to be expressed in a variety of human tissues (including colon, liver and lung parenchyme, mammary gland, and skin) and is overexpressed in certain cancer cell lines.9,10 Two cellular roles predominate in the literature regarding TIP47/perilipin-3 function: lipid droplet biogenesis and M6PR recycling.
In 1998, Diaz and Pfeffer identified a protein using a yeast 2-hybrid approach that interacted with the cytosolic tails of the M6PRs.6 M6PRs carry newly synthesized lysozomal enzymes from the Golgi apparatus to the endosomes where these enzymes are released. The M6PRs are subsequently returned to the Golgi to pick up additional hydrolases for transport. Diaz and Pfeffer provided evidence that this process of recycling was dependent upon both the protein they designated “tail-interacting protein of 47 kDa” (TIP47) and the small GTPase Rab9.6 They proposed that TIP47/perilipin-3 protected the M6PRs from degradation in the lysosome by interacting with the cytoplasmic tails of both the cation-dependent and cation-independent M6PRs and diverting them from the late endosome to the trans-Golgi network. Further studies by this group have implicated a host of other cellular proteins as participating in the recycling of the M6PRs.11-18
The second function ascribed to TIP47/perilipin-3 is a role in the biogenesis of lipid droplets. Lipid droplets are a newly recognized cellular organelle that were initially thought to be inert stores of excess lipids. However, increasing evidence suggests that they are also dynamically engaged in a number of other cellular functions, being functionally linked to the spliceosome and proteosome, playing a role in pathogen replication, and being involved in developmental processes (recently reviewed19). Based on the observation that TIP47/perilipin-3 shows homology to adipocyte differentiation-related protein (ADRP) and perilipin, some have suggested that TIP47/perilipin-3 performs a critical role in lipid biogenesis. Wolins et al. were the first to provide evidence that TIP47/perilipin-3 is associated with lipid droplets in HeLa cells.8 Subsequently, several studies using multiple techniques have shown association of TIP47/perilipin-3 with lipid droplets in multiple cell types.20-24
Some investigators view either M6PR recycling or lipid biogenesis as the predominant biological role for TIP47/perilipin-3, and others have provided evidence that the protein functions in both processes. Pauloin et al. have suggested that TIP47/perilipin-3 may be a so-called moonlighting protein, 25, a steadily increasing group of proteins that apparently perform multiple, disparate functions. 26 Supporting this moonlighting character of TIP47/perilipin-3 is the difference in intracellular localization as compared with those PAT family proteins involved solely in lipid biogenesis. TIP47/perilipin-3 appears to be distributed at multiple sites in the cell, including lipid droplets and the cytosol. 9,22,27
The structure of a C-terminal fragment of murine TIP47/perilipin-3 has been determined by X-ray crystallography.28 This truncated form comprises residues 191-437 encompassing the regions implicated in affecting M6PR binding but not those of Rab9 binding. The crystal structure reveals a distinctive L-shape, with the two arms of the L formed by a 4-helix bundle domain and an α/β domain with a hydrophobic cleft at the join. The hydrophobic cleft contains the residue Phe236 identified by Burguete et al. as important for cation-independent M6PR binding.29 This hydrophobic cleft has also been implicated in the binding of lipids, though not directly. It is postulated that another hydrophobic molecule binds in the cleft and mediates lipid binding to TIP47/perilipin-3.30
The C-terminal 4-helix bundle of TIP47/perilipin-3 has structural similarities to the N-terminal 4-helix bundle of apolipoprotein E (apoE); the root mean square deviation for Cα backbone atoms between these structures is 2.5 Å. ApoE has been shown to revise its helical orientation when bound to lipids, so that the four helices lie perpendicular to the lipid molecules.31 The C-terminal domain of TIP47/perilipin-3 has been shown to form lipid disks when incubated with liposomes in the same manner as apoE, and a similar rearrangement of the four helices has been proposed.21 If TIP47/perilipin-3 models the same domain organization and rearrangement as apoE, then it is the C-terminal domain that will coat lipid droplets, leaving the function of the N-terminal domain to be determined.
Additional biochemical and structural information on this enigmatic protein is needed to improve our understanding with respect to the cellular function or functions of TIP47/perilipin-3. We present here the results of studies aimed at characterizing the solution structure of the monomeric form of the human protein. We used small-angle X-ray scattering (SAXS), multiangle laser light scattering (MALLS), and circular dichroism (CD) to obtain information on the shape, oligomerization state, and secondary structure content of TIP47/perilipin-3.
MATERIALS AND METHODS
Cloning, overproduction, and purification of recombinant human TIP47/perilipin-3
The human TIP47/perilipin-3 gene was amplified from total cDNA generated from the CEM human T-lymphoblastoid cell line using primers designed to facilitate ligation-independent cloning into the vector pETHSUL.32 In addition to the full-length human TIP47/perilipin-3 (residues 1-434), three rationally designed truncation mutants were created, based on the work of Hickenbottom et al. and Sincock et al.28,33 pETHSUL vectors encoding N-terminal truncation mutants of human TIP47/perilipin-3 corresponding to amino acids 117-434, 152-434, and 187-434 were made using the ligation-independent cloning strategy. All vectors were verified by DNA sequencing.
Full-length human TIP47/perilipin-3 and the truncation mutants were overexpressed in Escherichia coli BL21 (DE3) Codon+RIL (Stratagene, La Jolla, CA) as N-terminal His6-SUMO (small ubiquitin-related modifier) protein fusions and purified using the same protocol. The culture was grown at 30 °C for 16 h using autoinducible media and harvested by centrifugation at 8600 × g for 7 min. Cell pellets were resuspended in 50 mM HEPES, pH 7.5, 500 mM KCl, 10 mM imidazole, 10% glycerol, and 5 mM β-mercaptoethanol (buffer A) plus an ethylenediamine tetra-acetic acid (EDTA)-free protease inhibitor cocktail (Roche, Basel, Switzerland). Cell lysis was achieved with two cycles of freeze-thaw employing liquid nitrogen, followed by a cycle of pressure shock (Rannie Laboratory homogenizer). The lysate was clarified by centrifugation at 48,000 × g for 35 min. The fusion protein was first purified on a nickel NTA (Qiagen, Valencia, CA) column preequilibrated in buffer A. The column was washed with 10-column volumes of buffer A and eluted with a 200-mL, 10-to 500-mM imidazole gradient, and 8-mL fractions were collected. Fractions were tested for protein using Bradford reagent and peak fractions were pooled. To the pooled fractions, 1 mM EDTA and 10 μg of a recombinant His6-tagged form of the catalytic domain (dtUD1) of the Saccharomyces cerevisiae SUMO hydrolase were added. Cleavage was allowed to proceed for 4 h at 18 °C. Following cleavage, the sample was dialyzed at 4 °C overnight against 2 L of 25 mM Tris-HCl pH 8.0 and 5 mM β-mercaptoethanol (buffer B). After dialysis, the SUMO hydrolase-catalyzed cleavage reaction was subjected to a second round of nickel affinity purification (in buffer B). This second column binds the His-tagged SUMO hydrolase, the cleaved His-SUMO, and any uncleaved protein. The flowthrough was collected containing the cleaved untagged TIP47/perilipin-3. The purified protein was then loaded onto a Q-sepharose column equilibrated in buffer B. The column was washed with 10-column volumes of buffer B and eluted with a 200-mL, 0- to 100-mM KCl gradient. Fractions were collected, concentrated, and further purified by size exclusion chromatography using a HiLoad 16/60 Superdex 75 or 200 prep grade column (Amersham Pharmacia Biotech, Piscataway, NJ) in a buffer containing 50 mM Tris-HCl, 250 mM NaCl, and 2.5 mM tris(2-carboxyethyl)phosphine (TCEP) pH 8.0, after which the samples were suitably pure for biophysical characterization.
Small-angle X-ray scattering
Sample preparation
Purified human TIP47/perilipin-3 and the truncation mutants TIP47/perilipin-3117-434, TIP47/perilipin-3152-434, and TIP47/perilipin-3187-434 were all dialyzed overnight against 50 mM Tris-HCl pH 8.0, 200 mM NaCl, and 5 mM TCEP at 4 °C to ensure complete solvent exchange. The final dialyzed samples had the following protein concentrations: TIP47/perilipin-3 (6.85 mg/mL), TIP47/perilipin-3117-434 (9.25 mg/mL), TIP47/perilipin-3152-434 (8.89 mg/mL), and TIP47/perilipin-3187-434 (8.22 mg/mL). Protein concentrations were determined after dialysis using the following extinction coefficients for A280nm (expressed as M−1 cm−1) calculated from the primary amino acid sequence using ProtParam34,35: TIP47/perilipin-3: 30,940; TIP47/perilipin-3117-434: 26,470; TIP47/perilipin-3152-434: 26,470; and TIP47/perilipin-3187-434: 26,470.
Data acquisition and analysis
SAXS data were acquired using an Anton-Paar SAXSess (Graz, Austria) with line collimation and as described by Jeffries et al.36 The instrument slit and integration lengths were set to 10 mm and beam profiles were recorded on an image plate detector. Scattering data were collected using the CCD detector; each sample was exposed for between 1 to 6 h with 10 integrations over 15-min blocks to improve counting statistics and enable checks for radiation damage. Samples were maintained at 20 °C.
Data reduction and analysis
Data were reduced to I(q) versus q (q = (4πsinθ)/β) where 2θ is the scattering angle between the incident and the scattered radiation, and β is the wavelength of the CuKα X-ray radiation (1.54 Å). Subtraction of the solvent scattering intensity was done with the program SAXSquant1D (Anton Paar) to yield the scattering from the protein. All data were placed on an absolute scale using the scattering from water as a standard.37 To identify potential protein aggregation and to estimate the forward scattering intensity at zero angle, I(0), and radius of gyration, Rg, of the proteins, Guinier analysis of the scattering data (corrected for instrument slit smearing) was done using the program PRIMUS.38 The programs GIFT and GNOM were used to calculate the indirect Fourier transform of the scattering to obtain the probable pair-wise distribution of interatomic distances within the scattering particle P(r). These programs account for the data smearing due to the slit geometry of the instrument. From the P(r) profile I(0), Rg, and the maximum linear dimension, Dmax, of the proteins were determined. The molecular mass of the scattering particles in each sample was calculated as previously described,37 using calculated values for the protein partial specific volume (ν) and contrast based on the amino acid sequences of the proteins (where Δρ = scattering density of the protein minus that of the supporting solvent), the P(r)-derived I(0) values, and experimental protein concentrations. The scattering length density (ρ) of TIP47/perilipin-3 and the components in the supporting SAXS buffer were calculated using the program MULCh.39 The partial specific volume (ν) and total volume of each protein was calculated from its primary amino acid sequence using NucProt.40 Estimates of the protein volumes were determined from the SAXS data using the method of Fischer et al.41 and were derived from the qmax-corrected scattering invariant of each construct calculated from the area under Kratky plots (I(q)q2 vs q) normalized to unit I(0) with corrections accounting for the finite measuring range for q (qmax = 0.3 Å−1; Suppl. Fig. 4.)
SAXS data were collected from each construct at four protein concentrations between 3 and 8 mg/mL. No concentration-dependent aggregation or interparticle interference effects that might bias the determined structural parameters were evident in the data.
Modeling
Ab initio shape restoration calculations for each TIP47/perilipin-3 truncation were performed using the P(r) results from GIFT as input to the program DAMMIF.42 Fifteen independent DAMMIF calculations were computed for each sample, and the resulting models were averaged and filtered using the program DAMAVER to generate a consensus envelope for each species. The mean normalized spatial discrepancies within each set of models were as follows: TIP47/perilipin-3: 0.815; TIP47/perilipin-3117-434: 0.77; TIP47/perilipin-3152-434: 0.815; and TIP47/perilipin-3187-434: 0.633, indicating a reasonable degree of similarity between each individual model within each set.43 The three TIP47/perilipin-3 models and the mouse crystal structure were aligned using the program SUPCOMB.44
Multiangle laser light scattering
MALLS data were obtained from purified full-length human TIP47/perilipin-3 and truncation mutants TIP47/perilipin-3117-434, TIP47/perilipin-3152-434, and TIP47/perilipin-3187-434. The MALLS system consisted of a Superdex 200 10/30 gel filtration column (Amersham) operated by an AKTA FPLC system (fast protein liquid chromatography; Amersham) feeding into a Wyatt Technology (Santa Barbara, CA) miniDAWN light scattering unit and a Wyatt Optilab DSP refractometer. The intrinsic Rayleigh scattering in toluene was used to calibrate the scattering to an absolute scale. The system was equilibrated with 5-column volumes of 50 mM Tris-HCl pH 8.0, 200 mM NaCl, and 5 mM TCEP, the same as the buffer in which the samples were prepared. For each of the four constructs, a 200-μL, 0.5-mg/mL sample was injected and the flow-through monitored. The relative molecular masses of each species were calculated assuming a uniform refractive index to concentration gradient (dn/dc) of 0.19 mL/g.
Circular dichroism spectropolarimetry
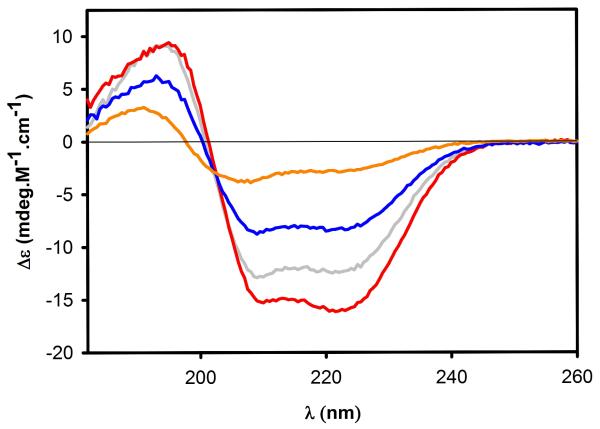
Protein samples were dialyzed against 20 mM Tris-HCl and 1 mM TCEP pH 8.0 at 4 °C for 18 h prior to analysis to ensure complete solvent exchange, at the following protein concentrations: TIP47/perilipin-3 (12.7 μM), TIP47/perilipin-3117-434 (28.1 μM), TIP47/perilipin-3152-434 (50.2 μM), and TIP47/perilipin-3187-434 (39.2 μM). Concentrations were determined after dialysis using the same extinction coefficients used to determine the protein concentrations for the SAXS samples. CD spectra for all TIP47/perilipin-3 constructs were performed on a Jasco J-720 spectropolarimeter (Jasco, Inc., Easton, MD). Spectra were monitored between 260 and 182 nm using a bandwidth of 0.5 nm, a response time of 1 second, and a scan speed of 20 nm/min. Each spectrum (Fig. 3) is an average of four scans. All data were recorded at 20 °C using a 0.1-mm path length cell. Spectra were processed using the CDPro suite of programs CONTIN, SELCON, and CDSSTR45 (protein basis sets 4 and 7) to determine the secondary structure content of all the TIP47/perilipin-3 constructs. For each sample, the background spectrum was subtracted from the protein spectrum.
Figure 3.
Far-UV spectra determining the secondary structure of full-length TIP47/perilipin-3 (gray line) and TIP47/perilipin-3 117-434 (red line), 152-434 (blue line), and 187-434 (orange line). Data were acquired over the range 182-260 nm and the plots have been scaled to molar protein concentration.
RESULTS AND DISCUSSION
The two important goals for this study were: (1) to resolve whether TIP47/perilipin-3 forms a stable oligomer, and if so of what size and what domain or region of the protein is responsible; and (2) to structurally characterize TIP47/perilipin-3 in order to define as completely as possible those domains or regions of the protein not present in the crystal structure from solution studies. Addressing these goals required the development of an innovative strategy for the production of highly pure protein preparations on a 1- to 10-mg scale.
Overproduction, purification, and isolation of monomeric recombinant full-length and N-terminally truncated human TIP47/perilipin-3 proteins
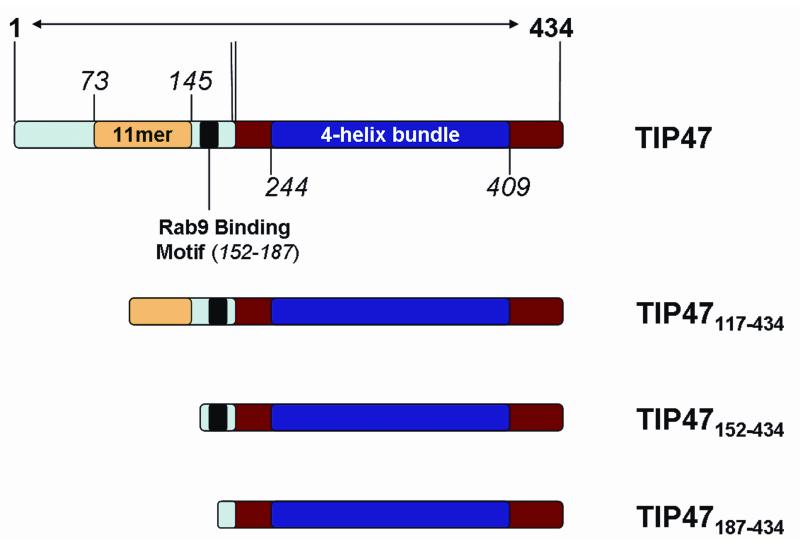
The X-ray crystal structure of an N-terminally truncated form of murine TIP47/perilipin-3 has been determined.28 This truncate, termed the PAT-C region (perilipin/ADRP/TIP47) (residues 191-437), has a structure that consists of an α/β domain of novel topology and a 4-helix bundle that resembles the low-density lipoprotein receptor binding domain of apoE.28 However, to date, no large-scale overexpression systems have been reported to overproduce milligram quantities of full-length human TIP47/perilipin-3 amenable to biophysical or structural studies. In an attempt to resolve this we created four bacterial TIP47/perilipin-3 constructs based on pETHSUL, an expression vector developed in the Loll laboratory.32 These vectors encode fusions in which the TIP47/perilipin-3 sequences are attached to an N-terminal His6-SUMO. Attachment of SUMO to the N-terminus of exogenous proteins is purported to dramatically enhance their expression in E. coli.46 Addition of the SUMO tag also affords a facile purification strategy: after the fusion protein is isolated by immobilized metal affinity chromatography (IMAC), the tag is removed by a His-tagged SUMO-specific protease, which allows for subtractive purification via a second IMAC column and the isolation of wild-type protein constructs without the inclusion or addition of affinity tags. The domain organization of human TIP47/perilipin-3 and the three N-terminally truncated mutants is shown in Figure 1. Structural domains include the 4-helix bundle and an α/β domain identified in the crystal structure of TIP47/perilipin-3 (residues 191-437 of mouse isoform) and an 11-mer repeat that is predicted to form a right-handed coiled-coil helical structure. The N-terminus of TIP47/perilipin-3 has been implicated in TIP47/perilipin-3 oligomerization, while the C-terminal sequence has been implicated in lipid binding.21 Using the pETHSUL expression system, and combining IMAC, ion exchange, and size exclusion chromatographies as outlined in Materials and Methods, we have isolated all of the TIP47/perilipin-3 constructs in purities greater than 90% (Suppl. Fig. 3) and in the following yields per liter of bacterial cell culture: TIP47/perilipin-3, 10 mg/L; TIP47/perilipin-3117-434, 31 mg/L; TIP47/perilipin-3152-434, 45 mg/L; and TIP47/perilipin-3187-434, 26 mg/L.
Figure 1.
Domain structure of full-length human TIP47/perilipin-3 and the N-terminal constructs generated for this study. The N-terminal regions are highlighted in light blue with yellow and black inserts indicating the 11-mer repeat and Rab9 binding regions, respectively. The C-terminal region corresponding to the mouse crystal structure is in red, with the 4-bundle helix region as a dark blue insert.
TIP47/perilipin-3 oligomerization state
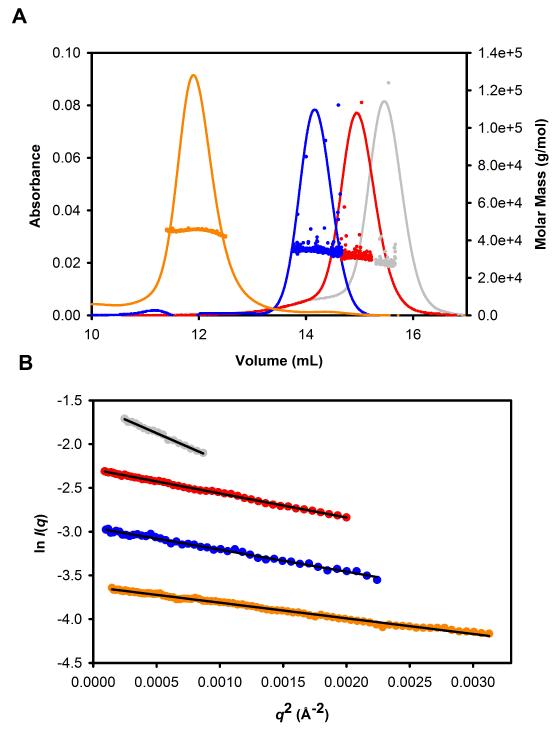
MALLS data were acquired for the full-length TIP47/perilipin-3 and the three truncation mutants (Fig. 2A). The MALLS data indicate that each of the three N-terminal truncations is monomeric (Fig. 2A), consistent with the linear Guinier plots of the SAXS data (Fig. 2B) and I(0)-dependent molecular mass analysis (Table I). This finding is also consistent with previous results from the Pfeffer group.33 However, in contrast to previous reports,33 in our hands full-length TIP47/perilipin-3 purified as two peaks after gel filtration. The major peak due to the lower-molecular-weight species is a monomer as shown by both MALLS and SAXS analysis. The additional peak, corresponding to a larger-molecular-weight species, was identified as a dimer in MALLS, but SAXS analysis suggested it was a mixture that included higher-molecular-weight species. As the SAXS experiments are performed at higher protein concentrations compared with MALLS (5-10 mg/mL vs 0.1 mg/mL), the full-length protein thus appears to show concentration-dependent self-association (data not shown).
Figure 2.
A: MALLS results for human TIP47/perilipin-3 and the truncation mutants. UV trace (lines) and MALLS experimental molar masses for full-length TIP47/perilipin-3 (gray), TIP47/perilipin-3117-434 (red), TIP47/perilipin-3152-434 (blue), and TIP47/perilipin-3187-434 (orange; see also Table I). B: Guinier plots from the SAXS data for the same protein constructs as in part A.
TABLE I.
Comparison of molecular mass values, Mr, calculated from primary sequences and those determined experimentally using MALLS or SAXS for full-length human TIP47/perilipin-3 and the truncation mutants. Predicted volumes based on primary amino acid sequences using the program NucProt with the volumes determined by SAXS from the scattering invariant corrected for the finite q-range measured.
|
Mr (calculated) |
Mr (MALLS) |
Mr (SAXS) |
Predicted Volume (NucProt) |
Corrected volumes (SAXS data) |
|
|---|---|---|---|---|---|
| Full-length human TIP47/perilipin-3 |
47,033 | 41,639 | 45,170 | 57,527 | 72,492 |
| TIP47/perilipin-3117-434 | 34,850 | 35,560 | 34,761 | 34,015 | 36,192 |
| TIP47/perilipin-3152-434 | 31,288 | 32,490 | 32,285 | 38,210 | 40,948 |
| TIP47/perilipin-3187-434 | 27,780 | 28,820 | 28,086 | 42,549 | 46,783 |
MALLS = multiangle laser light scattering; SAXS = small-angle X-ray scattering.
In concordance with previous reports on this protein, we found that the full-length TIP47/perilipin-3 protein has a propensity to oligomerize (Suppl. Figs. 1 and 2). Supplementary Figure 1 shows the time dependence of this oligomerization; the gel filtration trace immediately after the Q-sepharose column (black) shows two main peaks; peak 1 is the monomer (by SAXS and MALLS, see below and Fig. 2) and peak 2 has a molecular weight close to a dimer (by MALLS.) The same pooled Q-sepharose sample with a 2-day wait at 4 °C before applying to the gel filtration column (blue trace) shows that the dimer is the predominant species, and there also appear to be some degradation products. Supplementary Figure 2B shows MALLS traces for full-length TIP47/perilipin-3. This sample, from a poor-quality purification, highlights a concentration-dependent oligomerization, with molecular weights ranging from a dimer to nearly a tetramer. However, using the combination of low temperature and reducing agents within the purification buffers, we were able to circumvent this propensity of TIP47/perilipin-3 to oligomerize. The extreme sensitivity of this protein during the purification process may be one of the factors that stymied previous biophysical investigations.
Previous reports by Sincock et al.,33 working with recombinant full-length TIP47/perilipin-3, concluded that both the human and bovine proteins were hexameric. Possible explanations for the differences between this observation and our results include the use of slightly different protein constructs (native N-terminus vs His-tagged) and addition of reducing agents (β-mercaptoethanol or TCEP) in our purification buffer. Human TIP47/perilipin-3 contains three cysteines, two of which occur within the first 60 residues. As such, it is possible that the association detected by Sincock et al. is a consequence of oxidation of cysteine residues and disulfide formation. Consistent with this interpretation, we observed some tendency toward oligomerization in the full-length protein even under reducing conditions, whereas the three N-terminal truncation mutants are pure monomers.
Secondary structure differences between the N- and C-terminal regions of human TIP47/perilipin-3
We used CD spectroscopy to evaluate the secondary structure content of the monomeric full-length and the N-terminal truncation versions of human TIP47/perilipin-3. The CD data from the shortest human TIP47187-434 variant, which is homologous to the C-terminal region of the mouse crystal structure, indicates the highest percentage of α-helical content among the variants evaluated (~40% vs ~60% for the mouse isoform; Table II). As the N-terminal region of human TIP47 is lengthened, there appears to be a systematic decrease in the overall percentage of amino acids involved in forming α-helical structure and a concomitant increase in the percentage of amino acids forming α-strand, turns, or random coil (Table II). Based on these data, the helical content of human TIP47/perilipin-3 appears to be more concentrated toward to the C-terminal regions of the protein, suggesting that the N- and C-terminal regions are distinct structural modules at the level of secondary structure content. Currently, there are no published structural data relating to the N-terminus of TIP47/perilipin-3, though a previous report has proposed that the 11-mer repeats located in the N-terminal region of the protein (Figure 1) adopt α-helical structures.47 Our results suggest the full-length protein has a reduced α-helical content toward the N-terminus of the protein, contrary to sequence-to-structure predictions. However, in the case of α-synuclein, the 11-mer repeat portion of this protein (also predicted to be α-helical based on amino acid sequence-to-structure predictions) is not a stable helical entity, but only adopts a helical structure upon the binding of lipids or detergent micelles.48-52 Assessing whether the 11-mer repeat of TIP47/perilipin-3, as with α-synuclein, undergoes a conformational change that progresses towards an α-helical state on interacting with lipids is an intriguing possibility.
TABLE II.
Comparison of secondary structure estimates from the crystal structure of residues 197-437 of mouse TIP47/perilipin-3 (Protein Data Bank entry 1SZI) with those determined from circular dichroism measurements on the full-length human TIP47/perilipin-3 and the truncation mutants
| α-Helix % (approx. no. of residues) |
β-Strand % (approx. no. of residues) |
Turn % (approx. no. of residues) |
Random turn % (approx. no. of residues) |
|
|---|---|---|---|---|
| Mouse TIP47/perilipin-3197-437 crystal structure |
60 (150) | 5 (13) | N/D | N/D |
| TIP47/perilipin-3187-434 | 40 (100) | 13 (32) | 20 (49) | 28 (69) |
| TIP47/perilipin-3152-434 | 35 (98) | 17 (48) | 21 (59) | 28 (79) |
| TIP47/perilipin-3117-434 | 30 (94) | 18 (57) | 21 (67) | 31 (99) |
| Full-length human TIP47/perilipin-3 |
26 (113) | 20 (87) | 23 (100) | 31 (135) |
Full-length human TIP47/perilipin-3 adopts an extended conformation in solution
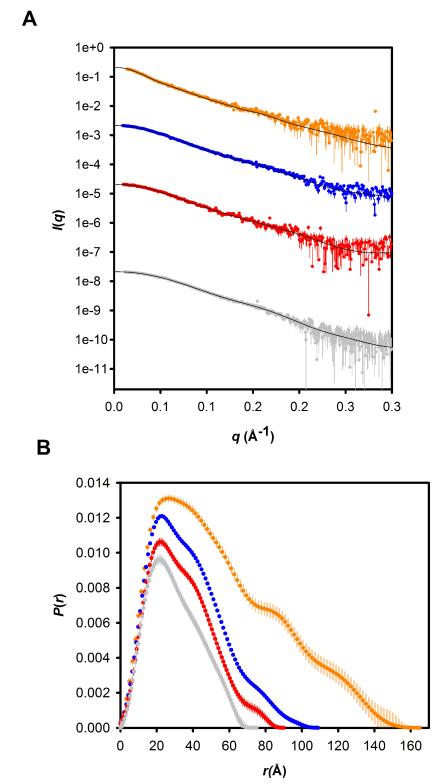
Next, we sought to extend the analysis of TIP47/perilipin-3 from secondary structure content to domain organization. We turned to SAXS, which can be used to probe the overall shape of monodisperse proteins in solution and in favorable circumstances provides information on domain dispositions within proteins. By carefully selecting size-exclusion eluates corresponding to “monomer-peaks,” we were able to analyze the shape of each TIP47/perilipin-3 construct as a free monomer in solution. The fact that the solutions contain monodisperse, monomeric forms was subsequently confirmed by molecular mass determinations from the forward scattering intensities I(0) derived from each construct’s respective SAXS dataset (Guinier analyses; Fig. 2 and Table I). The P(r) profiles calculated from the SAXS data (Fig. 4) show that the truncated TIP47/perilipin-3 variants as well as the full-length protein are highly elongated particles. Each P(r) profile peaks near 20 Å, and then tails toward zero systematically at increasing values of Rg and Dmax as the constructs incorporate more of the N-terminal regions of the protein (Fig. 4 and Table III). These observations indicate that as amino acids are added on to the C-terminal PAT-C domains, the structure becomes increasingly elongated via structural extensions in the direction of the long axis of the protein (as opposed to adopting a globular conformation). Furthermore, as the N-terminal regions extend into solution away from the C-terminal domain there may also be the accommodation of additional “twists-and-bends” along the length of the protein as suggested by the appearance of shoulders in the P(r) profile at mid-to long-range vector lengths (85 Å and 120 Å).
Figure 4.
A: Scattering data as I(q) versus q for human TIP47/perilipin-3 and the N-terminal truncation mutants. Full-length TIP47/perilipin-3 (gray, 6.85 mg/mL), TIP47/perilipin-3117-434 (red, 9.25 mg/mL), TIP47/perilipin-3152-434 (blue, 8.89 mg/mL), and TIP47/perilipin-3187-434 (orange, 8.22 mg/mL). The plots for TIP47/perilipin-3117-434, TIP47/perilipin-3152-434, and TIP47/perilipin-3187-434 have been arbitrarily shifted on the vertical axis for clarity. The solid lines are the fits to the data of the best-fit dummy atom models for each construct shown in Fig. 5. B: Corresponding P(r) profiles scaled to the ratio of the squares of the molecular weights of each protein.
TABLE III.
SAXS parameters for full-length human TIP47/perilipin-3 and the truncation mutants
| Full- length |
117-434 | 152-434 | 187-434 | |
|---|---|---|---|---|
| Structural parameters | ||||
| I(0) (cm−1) from P(r) | 0.2124 | 0.217 | 0.216 | 0.1795 |
| Rg (Å) from P(r) | 45.7 | 29.6 | 26.2 | 23.4 |
| Rg (Å) Guinier, qRg>1.3 | 43.5 ± 0.6 | 28.8 ± 0.04 | 27.5 ± 0.16 | 23.2 ± 0.07 |
| Dmax (Å) from P(r) | 165 ± 5 | 109 ± 5 | 90 ± 5 | 75 ± 5 |
| Molecular mass determination | ||||
| Protein concentration (g · cm−3) |
0.00685 | 0.00925 | 0.00889 | 0.00822 |
| Partial specific volume ν (cm3 · g−1) | 0.733 | 0.734 | 0.733 | 0.733 |
| Contrast 1010 · cm−2 | 2.890 | 2.875 | 2.908 | 2.884 |
| Mr from I(0) | 45,170 | 34,761 | 32,285 | 28,086 |
|
Mr from amino acid sequence |
47,033 | 34,850 | 31,288 | 27,780 |
SAXS = small-angle X-ray scattering.
Kratky plots of the scattering data (Suppl. Fig. 4) suggest that there may be a small increase in flexibility with increasing length of the protein TIP47/perilipin-3 constructs. The Kratky plot for the shortest version of the protein (corresponding to the C-terminal domain) goes toward zero at high q values as expected for a protein of defined shape, whereas the plots for longer constructs deviate from this path, showing a relatively small but nonzero asymptote indicating a degree of flexibility. (The Kratky plot of a flexible protein continues to rise with increasing q values, hence the conclusion in this case is that the flexibility is relatively limited.) The full-length protein appears to have more flexibility compared with the intermediate-length constructs. This observation is consistent with the average molecular volume calculations derived from the scattering invariant of the Kratky plot of the data (Suppl. Fig. 4). As residues are added to the C-terminal domain, the experimental average volumes also systematically increase over and above what is expected from the amino acid sequence (from 6% larger for TIP47/perilipin-3187-434 to 10% larger for TIP47/perilipin-3117-434 to ~26% larger for the full-length protein) while the mass values calculated from I(0) are as predicted from the sequence (Table I). The results thus suggest increasing relative flexibility with additional N-terminal sequence. This increase in flexibility upon addition of N-terminal sequences may be one of the reasons that only the relatively rigid C-terminal domain could be crystallized.
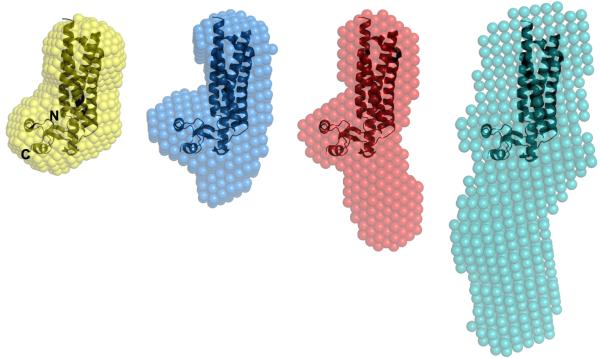
Ab initio shape restoration calculations for each TIP47/perilipin-3 construct provided models that could be compared with the crystal structure of the mouse homologue (Fig. 5). The TIP47/perilipin-3187-434 construct was designed based on the domain boundaries of the construct used for the mouse crystal structure.28 Accordingly, the overlay of the mouse crystal structure with the human TIP47/perilipin-3187-434 envelope in Figure 5 shows them to have very similar shapes. This is not surprising given that the mouse and human TIP47/perilipin-3 have 75% sequence identity. It is also possible to overlay the shapes of human TIP47/perilipin-3152-434 and TIP47/perilipin-3117-434 with the mouse crystal structure so as to highlight the extension of the extra mass from the C-terminal 4-helix bundle (Fig. 5). The PAT-C domain mouse crystal structure is represented as ribbons at the top of the restored molecular envelopes where it appears to superimpose best into the envelope; below the mouse crystal structure is the region of TIP47/perilipin-3 occupied by the N-terminal amino acids, which includes the 11-mer repeat. As the P(r) profiles indicate, the constructs become increasingly elongated as residues are added to the C-terminal domains so that the molecular envelope determined for the full-length protein has the most elongated shape in solution with a slight bend at one end. It must be noted that the normalized spatial discrepancy of the full-length protein was the highest of all of the molecular restorations (0.815), which indicates a variance in shape across the individual solutions used to generate the molecular envelope shown in Figure 5. The asymmetric shape of full-length TIP47/perilipin-3, combined with the indications that it has the largest degree of flexibility, is the likely cause of these variations across the individual solutions. The molecular envelope in Fig. 5 does not represent a single rigid structure but rather is an average, slightly expanded volume representing the protein structural ensemble.
Figure 5.
Alignment of the mouse TIP47/perilipin-3 PAT-C domain crystal structure (PDB 1SZI; ribbons) spatially positioned within the consensus shape-models (spheres) derived from the SAXS data for human full-length TIP47/perilipin-3 (gray); TIP47/perilipin-3117-434 (red);TIP47/perilipin-3152-434 (blue) and; TIP47/perilipin-3187-434 (orange). The mouse homologue spatially superimposes into the molecular shape of the shortest human truncation mutant (TIP47/perilipin-3187-434) that spans the C-terminal domains of the protein. The space occupied by regions encompassing the N-terminal half of human TIP47/perilipin-3 appears to extend into solution away from the C-terminal domains.
In summary, the SAXS data indicate that monomeric full-length human TIP47/perilipin-3 adopts a highly elongated shape in solution, with some flexibility introduced with the addition of the N-terminal sequence. Further, there is a spatial separation of the N- and C-terminal regions in monomeric human TIP47/perilipin-3 by the α/β domain, which opens the interesting possibility that the N- and C-terminal domains may act as distinct “functional modules” when associating with either proteins or lipid droplets.
Supplementary Material
Supplementary Figure 1 Gel filtration trace of full-length TIP47/perilipin-3 immediately after (black trace) and 2 days after (blue trace) the Q-sepharose column. Peak 1 represents the monomer, which when rerun remains as a monomer and is the sample used for all the experiments (see Supplementary Figure 2).
Supplementary Figure 2 A: MALLS traces of full-length TIP47/perilipin-3; peak 1 from Supplementary Figure 1 (black trace) and peak 2 from Supplementary Figure 1 (blue trace). Molecular weights were 41,639 Da for peak 1 and 91,290 Da for peak 2. B: TIP47/perilipin-3 full-length UV trace (lines) and MALLS experimental molar masses for TIP47/perilipin-3 0.25 mg/mL (gray), 0.5 mg/mL (red), 2 mg/mL (blue), and 7 mg/mL (orange). Molecular weights determined for these are, respectively, 94,720; 95,410; 121,900; and 182,200 Da. Traces have been normalized to account for differing protein concentrations.
Supplementary Figure 3 SDS-PAGE (sodium dodecyl sulfate polyacrylamide gel electrophoresis) analysis of recombinant TIP47/perilipin-3. Lane 1: TIP47/perilipin-3117-434 after SUMO-hydrolase cleavage. Lane 2: TIP47/perilipin-3117-434 after 2nd IMAC column. Lanes 3-9: TIP47/perilipin-3117-434 fractions from a Q-sepharose column. This purity is representative of all constructs.
Supplementary Figure 4 Kratky plots of human TIP47/perilipin-3 (gray), TIP47/perilipin-3117-434 (red), TIP47/perilipin-3152-434 (blue), and TIP47/perilipin-3187-434 (orange). Scattering data and smoothed I(q) vs q profiles (from GIFT; continuous lines) were normalized to unit I(0) prior to transformation as per the method of Fischer et al.41 in order to obtain the qmax-corrected scattering invariant (area under the Kratky plots to 0.3 Å−1) from which the volume estimates of the scattering particles were determined.
Acknowledgments
The authors are grateful to Philippa Stokes (School of Molecular Bioscience, University of Sydney) for help with data collection for the circular dichroism experiments and to Diana Winters (Academic Publishing Services, Drexel University College of Medicine) for proofreading the manuscript.
Grant support: R03AI078790-01A1, P50 GM082545.
References
- 1.Kimmel AR, Brasaemle DL, McAndrews-Hill M, Sztalryd C, Londos C. Adoption of PERILIPIN as a unifying nomenclature for the mammalian PAT-family of intracellular lipid storage droplet proteins. J Lipid Res. 2010;51:468–471. doi: 10.1194/jlr.R000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brasaemle DL. Thematic review series: adipocyte biology. The perilipin family of structural lipid droplet proteins: stabilization of lipid droplets and control of lipolysis. J Lipid Res. 2007;48:2547–2559. doi: 10.1194/jlr.R700014-JLR200. [DOI] [PubMed] [Google Scholar]
- 3.Bauby H, Lopez-Verges S, Hoeffel G, Delcroix-Genete D, Janvier K, Mammano F, Hosmalin A, Berlioz-Torrent C. TIP47 is required for the production of infectious HIV-1 particles from primary macrophages. Traffic. 2010;11:455–467. doi: 10.1111/j.1600-0854.2010.01036.x. [DOI] [PubMed] [Google Scholar]
- 4.Bohn H, Kraus W, Winckler W. Purification and characterization of two new soluble placental tissue proteins (PP13 and PP17) Oncodev Biol Med. 1983;4:343–350. [PubMed] [Google Scholar]
- 5.Chen Y, Honeychurch KM, Yang G, Byrd CM, Harver C, Hruby DE, Jordan R. Vaccinia virus p37 interacts with host proteins associated with LE-derived transport vesicle biogenesis. Virol J. 2009;6:44. doi: 10.1186/1743-422X-6-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Diaz E, Pfeffer SR. TIP47: a cargo selection device for mannose 6-phosphate receptor trafficking. Cell. 1998;93:433–443. doi: 10.1016/s0092-8674(00)81171-x. [DOI] [PubMed] [Google Scholar]
- 7.Tsuiki E, Fujita A, Ohsaki Y, Cheng J, Irie T, Yoshikawa K, Senoo H, Mishima K, Kitaoka T, Fujimoto T. All-trans-retinol generated by rhodopsin photobleaching induces rapid recruitment of TIP47 to lipid droplets in the retinal pigment epithelium. Invest Ophthalmol Vis Sci. 2007;48:2858–2867. doi: 10.1167/iovs.06-0768. [DOI] [PubMed] [Google Scholar]
- 8.Wolins NE, Rubin B, Brasaemle DL. TIP47 associates with lipid droplets. J Biol Chem. 2001;276:5101–5108. doi: 10.1074/jbc.M006775200. [DOI] [PubMed] [Google Scholar]
- 9.Straub BK, Herpel E, Singer S, Zimbelmann R, Breuhahn K, Macher-Goeppinger S, Warth A, Lehmann-Koch J, Longerich T, Heid H, Schirmacher P. Lipid droplet-associated PAT-proteins show frequent and differential expression in neoplastic steatogenesis. Mod Pathol. 2010;23:480–492. doi: 10.1038/modpathol.2009.191. [DOI] [PubMed] [Google Scholar]
- 10.Than GN, Turoczy T, Sumegi B, Than NG, Bellyei S, Bohn H, Szekeres G. Overexpression of placental tissue protein 17b/TIP47 in cervical dysplasias and cervical carcinoma. Anticancer Res. 2001;21:639–642. [PubMed] [Google Scholar]
- 11.Diaz E, Schimmoller F, Pfeffer SR. A novel Rab9 effector required for endosome-to-TGN transport. J Cell Biol. 1997;138:283–290. doi: 10.1083/jcb.138.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Derby MC, Lieu ZZ, Brown D, Stow JL, Goud B, Gleeson PA. The trans-Golgi network golgin, GCC185, is required for endosome-to-Golgi transport and maintenance of Golgi structure. Traffic. 2007;8:758–773. doi: 10.1111/j.1600-0854.2007.00563.x. [DOI] [PubMed] [Google Scholar]
- 13.Reddy JV, Burguete AS, Sridevi K, Ganley IG, Nottingham RM, Pfeffer SR. A functional role for the GCC185 golgin in mannose 6-phosphate receptor recycling. Mol Biol Cell. 2006;17:4353–4363. doi: 10.1091/mbc.E06-02-0153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ganley IG, Espinosa E, Pfeffer SR. A syntaxin 10-SNARE complex distinguishes two distinct transport routes from endosomes to the trans-Golgi in human cells. J Cell Biol. 2008;180:159–172. doi: 10.1083/jcb.200707136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burguete AS, Fenn TD, Brunger AT, Pfeffer SR. Rab and Arl GTPase family members cooperate in the localization of the golgin GCC185. Cell. 2008;132:286–298. doi: 10.1016/j.cell.2007.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Lartigue J, Polson H, Feldman M, Shokat K, Tooze SA, Urbe S, Clague MJ. PIKfyve regulation of endosome-linked pathways. Traffic. 2009;10:883–893. doi: 10.1111/j.1600-0854.2009.00915.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Espinosa EJ, Calero M, Sridevi K, Pfeffer SR. RhoBTB3: a Rho GTPase-family ATPase required for endosome to Golgi transport. Cell. 2009;137:938–948. doi: 10.1016/j.cell.2009.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doray B, Ghosh P, Griffith J, Geuze HJ, Kornfeld S. Cooperation of GGAs and AP-1 in packaging MPRs at the trans-Golgi network. Science. 2002;297:1700–1703. doi: 10.1126/science.1075327. [DOI] [PubMed] [Google Scholar]
- 19.Fujimoto T, Parton RG. Not just fat: the structure and function of the lipid droplet. Cold Spring Harb Perspect Biol. 2011:3. doi: 10.1101/cshperspect.a004838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bickel PE, Tansey JT, Welte MA. PAT proteins, an ancient family of lipid droplet proteins that regulate cellular lipid stores. Biochim Biophys Acta. 2009;1791:419–440. doi: 10.1016/j.bbalip.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bulankina AV, Deggerich A, Wenzel D, Mutenda K, Wittmann JG, Rudolph MG, Burger KN, Honing S. TIP47 functions in the biogenesis of lipid droplets. J Cell Biol. 2009;185:641–655. doi: 10.1083/jcb.200812042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miura S, Gan JW, Brzostowski J, Parisi MJ, Schultz CJ, Londos C, Oliver B, Kimmel AR. Functional conservation for lipid storage droplet association among Perilipin, ADRP, and TIP47 (PAT)-related proteins in mammals, Drosophila, and Dictyostelium. J Biol Chem. 2002;277:32253–32257. doi: 10.1074/jbc.M204410200. [DOI] [PubMed] [Google Scholar]
- 23.Robenek H, Robenek MJ, Buers I, Lorkowski S, Hofnagel O, Troyer D, Severs NJ. Lipid droplets gain PAT family proteins by interaction with specialized plasma membrane domains. J Biol Chem. 2005;280:26330–26338. doi: 10.1074/jbc.M413312200. [DOI] [PubMed] [Google Scholar]
- 24.Robenek H, Robenek MJ, Troyer D. PAT family proteins pervade lipid droplet cores. J Lipid Res. 2005;46:1331–1338. doi: 10.1194/jlr.M400323-JLR200. [DOI] [PubMed] [Google Scholar]
- 25.Pauloin A, Ollivier-Bousquet M, Chanat E. [The double-play of PP17/TIP47] Med Sci (Paris) 2004;20:1020–1025. doi: 10.1051/medsci/200420111020. [DOI] [PubMed] [Google Scholar]
- 26.Huberts DH, van der Klei IJ. Moonlighting proteins: an intriguing mode of multitasking. Biochim Biophys Acta. 2010;1803:520–525. doi: 10.1016/j.bbamcr.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 27.Barbero P, Buell E, Zulley S, Pfeffer SR. TIP47 is not a component of lipid droplets. J Biol Chem. 2001;276:24348–24351. doi: 10.1074/jbc.M102468200. [DOI] [PubMed] [Google Scholar]
- 28.Hickenbottom SJ, Kimmel AR, Londos C, Hurley JH. Structure of a lipid droplet protein; the PAT family member TIP47. Structure. 2004;12:1199–1207. doi: 10.1016/j.str.2004.04.021. [DOI] [PubMed] [Google Scholar]
- 29.Burguete AS, Harbury PB, Pfeffer SR. In vitro selection and prediction of TIP47 protein-interaction interfaces. Nat Methods. 2004;1:55–60. doi: 10.1038/nmeth702. [DOI] [PubMed] [Google Scholar]
- 30.Ohsaki Y, Maeda T, Maeda M, Tauchi-Sato K, Fujimoto T. Recruitment of TIP47 to lipid droplets is controlled by the putative hydrophobic cleft. Biochem Biophys Res Commun. 2006;347:279–287. doi: 10.1016/j.bbrc.2006.06.074. [DOI] [PubMed] [Google Scholar]
- 31.Raussens V, Fisher CA, Goormaghtigh E, Ryan RO, Ruysschaert JM. The low density lipoprotein receptor active conformation of apolipoprotein E. Helix organization in n-terminal domain-phospholipid disc particles. J Biol Chem. 1998;273:25825–25830. doi: 10.1074/jbc.273.40.25825. [DOI] [PubMed] [Google Scholar]
- 32.Weeks SD, Drinker M, Loll PJ. Ligation independent cloning vectors for expression of SUMO fusions. Protein Expr Purif. 2007;53:40–50. doi: 10.1016/j.pep.2006.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sincock PM, Ganley IG, Krise JP, Diederichs S, Sivars U, O’Connor B, Ding L, Pfeffer SR. Self-assembly is important for TIP47 function in mannose 6-phosphate receptor transport. Traffic. 2003;4:18–25. doi: 10.1034/j.1600-0854.2003.40104.x. [DOI] [PubMed] [Google Scholar]
- 34.Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003;31:3784–3788. doi: 10.1093/nar/gkg563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wilkins MR, Gasteiger E, Bairoch A, Sanchez JC, Williams KL, Appel RD, Hochstrasser DF. Protein identification and analysis tools in the ExPASy server. Methods Mol Biol. 1999;112:531–552. doi: 10.1385/1-59259-584-7:531. [DOI] [PubMed] [Google Scholar]
- 36.Jeffries CM, Whitten AE, Harris SP, Trewhella J. Small-angle X-ray scattering reveals the N-terminal domain organization of cardiac myosin binding protein C. J Mol Biol. 2008;377:1186–1199. doi: 10.1016/j.jmb.2008.01.080. [DOI] [PubMed] [Google Scholar]
- 37.Orthaber D, Bergmann A, Glatter O. SAXS experiments on absolute scale with Kratky systems using water as a secondary standard. J Appl Crystallogr. 2000;33:218–225. [Google Scholar]
- 38.Konarev PV, Volkov VV, Sokolova AV, Koch MHJ, Svergun DI. PRIMUS: a Windows PC-based system for small-angle scattering data analysis. J Appl Crystallogr. 2003;36:1277–1282. [Google Scholar]
- 39.Whitten AE, Cai SZ, Trewhella J. MULCh: modules for the analysis of small-angle neutron contrast variation data from biomolecular assemblies. J Appl Crystallogr. 2008;41:222–226. [Google Scholar]
- 40.Voss NR, Gerstein M. Calculation of standard atomic volumes for RNA and comparison with proteins: RNA is packed more tightly. J Mol Biol. 2005;346:477–492. doi: 10.1016/j.jmb.2004.11.072. [DOI] [PubMed] [Google Scholar]
- 41.Fischer H, Neto MD, Napolitano HB, Polikarpov I, Craievich AF. Determination of the molecular weight of proteins in solution from a single small-angle X-ray scattering measurement on a relative scale. J Appl Crystallogr. 2010;43:101–109. [Google Scholar]
- 42.Franke D, Svergun DI. DAMMIF, a program for rapid ab-initio shape determination in small-angle scattering. J Appl Crystallogr. 2009;42:342–346. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Volkov VV, Svergun DI. Uniqueness of ab initio shape determination in small-angle scattering. J Appl Crystallogr. 2003;36:860–864. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kozin MB, Svergun DI. Automated matching of high- and low-resolution structural models. J Appl Crystallogr. 2001;34:33–41. [Google Scholar]
- 45.Sreerama N, Woody RW. Estimation of protein secondary structure from circular dichroism spectra: Comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set. Anal Biochem. 2000;287:252–260. doi: 10.1006/abio.2000.4880. [DOI] [PubMed] [Google Scholar]
- 46.Malakhov MP, Mattern MR, Malakhova OA, Drinker M, Weeks SD, Butt TR. SUMO fusions and SUMO-specific protease for efficient expression and purification of proteins. J Struct Funct Genomics. 2004;5:75–86. doi: 10.1023/B:JSFG.0000029237.70316.52. [DOI] [PubMed] [Google Scholar]
- 47.Bussell R, Jr, Eliezer D. A structural and functional role for 11-mer repeats in alpha-synuclein and other exchangeable lipid binding proteins. J Mol Biol. 2003;329:763–778. doi: 10.1016/s0022-2836(03)00520-5. [DOI] [PubMed] [Google Scholar]
- 48.Davidson WS, Jonas A, Clayton DF, George JM. Stabilization of α-synuclein secondary structure upon binding to synthetic membranes. J Biol Chem. 1998;273:9443–9449. doi: 10.1074/jbc.273.16.9443. [DOI] [PubMed] [Google Scholar]
- 49.Chandra S, Chen XC, Rizo J, Jahn R, Sudhof TC. A broken α-helix in folded α-synuclein. J Biol Chem. 2003;278:15313–15318. doi: 10.1074/jbc.M213128200. [DOI] [PubMed] [Google Scholar]
- 50.Eliezer D, Kutluay E, Bussell R, Browne G. Conformational properties of alpha-synuclein in its free and lipid-associated states. J Mol Biol. 2001;307:1061–1073. doi: 10.1006/jmbi.2001.4538. [DOI] [PubMed] [Google Scholar]
- 51.Bartels T, Ahlstrom LS, Leftin A, Kamp F, Haas C, Brown MF, Beyer K. The N-terminus of the intrinsically disordered protein a-synuclein triggers membrane binding and helix folding. Biophys J. 2010;99:2116–2124. doi: 10.1016/j.bpj.2010.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weinreb PH, Zhen WG, Poon AW, Conway KA, Lansbury PT. NACP, a protein implicated in Alzheimer’s disease and learning, is natively unfolded. Biochemistry. 1996;35:13709–13715. doi: 10.1021/bi961799n. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1 Gel filtration trace of full-length TIP47/perilipin-3 immediately after (black trace) and 2 days after (blue trace) the Q-sepharose column. Peak 1 represents the monomer, which when rerun remains as a monomer and is the sample used for all the experiments (see Supplementary Figure 2).
Supplementary Figure 2 A: MALLS traces of full-length TIP47/perilipin-3; peak 1 from Supplementary Figure 1 (black trace) and peak 2 from Supplementary Figure 1 (blue trace). Molecular weights were 41,639 Da for peak 1 and 91,290 Da for peak 2. B: TIP47/perilipin-3 full-length UV trace (lines) and MALLS experimental molar masses for TIP47/perilipin-3 0.25 mg/mL (gray), 0.5 mg/mL (red), 2 mg/mL (blue), and 7 mg/mL (orange). Molecular weights determined for these are, respectively, 94,720; 95,410; 121,900; and 182,200 Da. Traces have been normalized to account for differing protein concentrations.
Supplementary Figure 3 SDS-PAGE (sodium dodecyl sulfate polyacrylamide gel electrophoresis) analysis of recombinant TIP47/perilipin-3. Lane 1: TIP47/perilipin-3117-434 after SUMO-hydrolase cleavage. Lane 2: TIP47/perilipin-3117-434 after 2nd IMAC column. Lanes 3-9: TIP47/perilipin-3117-434 fractions from a Q-sepharose column. This purity is representative of all constructs.
Supplementary Figure 4 Kratky plots of human TIP47/perilipin-3 (gray), TIP47/perilipin-3117-434 (red), TIP47/perilipin-3152-434 (blue), and TIP47/perilipin-3187-434 (orange). Scattering data and smoothed I(q) vs q profiles (from GIFT; continuous lines) were normalized to unit I(0) prior to transformation as per the method of Fischer et al.41 in order to obtain the qmax-corrected scattering invariant (area under the Kratky plots to 0.3 Å−1) from which the volume estimates of the scattering particles were determined.