In many organisms, differentiation of the sex chromosome complement resulted in the coordinated regulation of genes on whole chromosomes to equalize gene expression between the sexes. In mammals, X inactivation evolved to restore equal expression of X-linked genes in males and females (1). Although X inactivation consists in the general repression of most genes on the X, some genes escape inactivation (reviewed in ref. 2). Recent advances in the Human Genome Project now allow the inactivation status of many X-linked genes to be systematically studied. In this issue of PNAS, Carrel et al. (3) report such a systematic analysis. Their data are so extensive that they are summarized in the paper but are fully accessible only as a file on the World Wide Web (www.pnas.org/supplementary.shtml).
According to Carrel et al. (3), “escapees” (genes that escape X inactivation) are not rare: conservatively, they number as much as 19%, or about one-fifth, of the genes on the human X. Another striking finding of Carrel et al. (3) is that the distribution of escapees and of genes subject to X inactivation is nonrandom. Many more escapees are located on the short arm of the human X (21%) than on the long arm (3%). The origin of these patterns of expression and organization can be tied to the evolutionary history of the sex chromosomes. The existence of these patterns also has implications for the regulation and role of X chromosomal genes in human disease.
It is useful to begin by placing Carrel et al.’s findings in the context of our understanding of sex chromosome evolution. Ohno (4) proposed that the sex chromosomes derived from a homomorphic autosome-like pair of chromosomes. Determination of the male sex by a gene (SRY) on the Y resulted in inhibition of recombination between the sex chromosomes. Decay of the Y and accumulation of genes advantageous to males close to the male determinant resulted in dramatic differences in size and gene content between the sex chromosomes (5, 6). Extensive gene loss from the Y provided a strong impetus for X inactivation (Fig. 1). Expression from a single allele on the active X in both sexes would, however, have led to an expression imbalance relative to biallelic autosomal genes. Hence, up-regulation of genes on the active X likely evolved together with—or before—X inactivation (Fig. 1) (4, 7).
Figure 1.
Dosage of genes on the sex chromosomes in relation to biallelically expressed autosomal genes (blue). In females, genes on the X can either be subject to inactivation on the inactive X (white) and up-regulated on the active X (red), or they can escape (pink). In males, genes on the X are either up-regulated (red) or not (pink), and genes on the Y either are lost, are differentiated to acquire a male-specific function (green), or persist as functionally equivalent Y partners in X/Y gene pairs (pink).
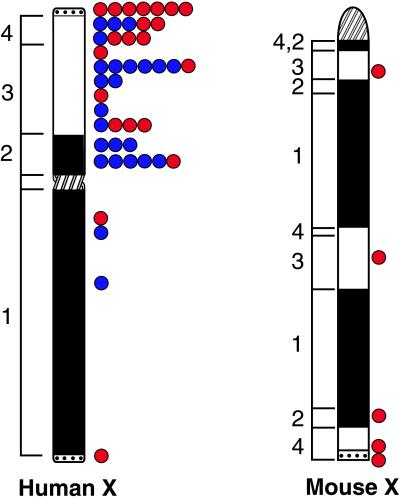
The ancestral sex chromosomes of mammals were probably smaller than the present-day eutherian X chromosomes. Graves (8) has suggested that most of the short arm of the human X is a recent addition to an ancestral chromosome. This ancestral region [X-conserved-region (XCR)] is also present in marsupials and monotremes, whereas the added region [X-added-region (XAR)] is present only in eutherians (Fig. 2). An interesting new study by Lahn and Page (9) defines four evolutionary strata on the human X chromosome on the basis of sequence divergence between X/Y genes. Thus at least four events (likely inversions of the Y) must have shaped the human sex chromosomes. The oldest strata, 1 and 2, correspond to the XCR, and the two newest strata, 3 and 4, to the XAR. The three most recent strata are all on the X short arm, precisely where the majority of escapees are located (Fig. 2)
Figure 2.
Ideograms of the human and mouse X chromosomes indicating the position of the centromere (crosshatched), of the XCR (X-conserved region, filled), of the XAR (X-added region, unfilled), and of the pseudoautosomal region (PAR), dotted (8). (Left) The approximate position of evolutionary strata, 1, 2, 3, and 4, defined by Lahn and Page (9). The position of escapees is indicated (Right) with the X/Y genes indicated in red (genes with functional and nonfunctional Y partners included) and the X-only escapees in blue.
The process of incorporation of genes into the X inactivation/up-regulation system within each stratum was likely progressive. In fact, it may have occurred on a gene-by-gene basis, with the Y gene being first lost or differentiated to acquire a male-specific function. Indeed, there is no known example of a gene that is subject to X inactivation but has not lost its functionally equivalent Y partner. Thus it seems that degeneration of the Y took place before the acquisition of X inactivation (9, 10). Studies in marsupials provide some information about acquisition of X inactivation. Marsupial X inactivation is incomplete and unstable, perhaps because of the absence of locking mechanisms (e.g., methylation), whereas genes on eutherian inactive Xs (except for escapees) are usually stably inactivated and methylated at their CpG islands (reviewed in refs. 11, 12). Histone deacetylation appears to be a more ancestral mechanism found in both eutherian and marsupial X inactivation. Although the marsupial X-inactive-specific-transcript gene (XIST) has not been identified, its hypothetical location lies in the XCR, in stratum 1. Thus it appears that some primitive form of X inactivation may have evolved before the divergence of eutherians and marsupials, and the XAR may have been translocated to an ancestral chromosome already regulated by X inactivation.
The escapees identified by Carrel et al. (3) in the short arm of the human X are likely remnants of incomplete incorporation of individual genes into the X inactivation system but could also result in part from incomplete inactivation spreading into regions added to an ancestral X. Whether X inactivation spreading occurred or not in the XAR, it is interesting to compare the results of Carrel et al. (3) with findings in X/autosome translocations. In such translocations, spreading is often incomplete, and many of the autosomal genes attached to the inactive X escape in a discontinuous fashion (13). Spreading, in cases of unbalanced X/autosome translocations, provides partial correction of the trisomic state and thus would be favorable. Yet, as in the study of Carrel et al. (3), gene inactivation is discontinuous and many genes escape. Thus a selection for corrected gene dosage may not operate on all genes, either because dosage is unimportant for some genes or because they cannot be stably inactivated.
One would expect that the high density of escapees on the X short arm would be accompanied by a high density of remaining X/Y gene pairs in this region. In Fig. 2, we have indicated the position of X/Y genes among the escapees, and indeed the density of X/Y gene pairs is higher on the short arm, which includes the three newest strata defined by Lahn and Page (9). The most distal short arm region is the pseudoautosomal region 1 (PAR1) where pairing occurs between the sex chromosomes. All genes that could be assayed in the PAR1 escape X inactivation, as expected for genes equally expressed from the X and Y. Another small pseudoautosomal region 2, located at the extremity of the long arms of the human sex chromosomes, represents a recent addition that contains one escapee and one gene that, surprisingly, is inactivated on both the X and the Y (14).
Functional equivalence, with biallelic expression in both sexes, has been observed for some nonpseudoautosomal X/Y genes (15, 16). Persistence of functional X/Y gene pairs in evolutionary distant species could be explained by their resistance to dosage changes. Other X/Y gene pairs offer glimpses in the evolutionary reduction of the Y and the differentiation of surviving Y genes into male-specific genes. Once the Y partner has been lost or has acquired a testis-specific function, one would expect the X gene to be incorporated in the X inactivation/up-regulation systems (Fig. 1). However, X inactivation often lags behind, which could explain at least some persistent escapees that have lost their Y homologue (Fig. 1 and 2). Dosage may not be important for such genes or, alternatively, dosage may be important for a sex-specific function. For example, some escapees may require higher expression in females for specific ovarian functions (Fig. 1).
Jegalian and Page (10) have shown that X/Y gene pair differentiation proceeded independently in different evolutionary lineages. Comparative analyses of the human and mouse X chromosomes provide interesting clues about evolution of the sex chromosomes and escape from inactivation. The evolutionary makeup of the human X appears relatively simple, with at least four evolutionary strata preserved in order from the extremity of the long arm to the tip of the short arm where the pseudoautosomal region 1 is located (Fig. 2) (8, 9). Many mammalian X chromosomes appear to have this basic structure (17). In contrast, the mouse X has been scrambled, with its centromere now located at one extremity and regions of conservation intermixed with added regions (Fig. 2) (18).
Mice seem to have fewer escapees, although many fewer genes have thus far been assayed in the rigorous fashion of Carrel et al. (3) (Fig. 2). Until such a systematic study is undertaken, it is difficult to know whether the mouse X is truly escapee poor. One indication that there may be fewer mouse escapees comes from XO mice, who have a less severe phenotype than do 45,X Turner females (reviewed in ref. 19). Interestingly, at least four pairs of differentiated X/Y genes with a testis-specific Y partner and an X partner subject to X inactivation have been found in mice, whereas only one such X/Y gene pair has been described in humans. Thus the mouse X chromosome may be further along in the evolution of X/Y gene pairs.
Differences in X chromosome structure between human and mouse may have led to evolutionary differences in X inactivation patterns (Fig. 2). In humans, but not in mice, the X-inactivation center that contains XIST is separated from the short arm (where the majority of escapees are located) by the centromere. Duthie et al. (20) recently examined rodent X chromosomes containing a large amount of heterochromatin through which the coating of Xist RNA did not proceed. In addition, the centromere of the mouse X was not coated with Xist. In contrast, in mouse X/autosome translocations and in transgenic mice containing Xist on an autosome, Xist RNA spread into the autosome (20, 21). Unfortunately, it has not been possible to study the precise distribution of XIST RNA on the human X or in X/autosome translocations, because this RNA does not remain associated with human metaphase chromosomes. Could centromeric heterochromatin provide a barrier between long and short arms of the human X, beyond which inactivation is incomplete, or is the high density of escapees on the X short arm simply related to the age of the strata? Extension of the approach of Carrel et al. (3) to study additional human and mouse genes will help sort out the role of evolutionary changes in chromosome structure between species.
The mechanisms by which genes escape X inactivation remain elusive. Sequence analyses of the promoter region of human ZFX (escapee) and mouse Zfx (subject to X inactivation) revealed remarkable conservation between the genes (22). We previously showed that an escapee can be completely turned off on the inactive X in embryonic cells and thus is susceptible to molecular changes associated with X inactivation (23). These findings suggest that maintenance and/or cell selection play an important role in establishing escape patterns during development. Carrel et al. (3) report that several of the human escapees have heterogeneous patterns of inactivation, i.e., they are expressed from the inactive X in some hybrids but not in others. Although these patterns may partly result from the instability of X inactivation in hybrid cell lines (reviewed in ref. 11), variable escape patterns have been confirmed in vivo (3, 24). Thus, analogous to imprinted genes, there may be tissue-specific patterns of escape in addition to developmentally regulated patterns.
Whether the control of X inactivation maintenance is at the level of individual genes or groups of genes within chromatin domains has not been established. A large domain of contiguous escapees was found on the very proximal short arm of the human X (25). The nearly complete sequence of the human X chromosome will allow one to refine the distribution of genes that escape. Carrel et al. plan to keep updating their list, which will continue to be available on the World Wide Web. Ultimately, the identification of matrix attachment regions will be critical to evaluate the role of chromatin domains in regulating expression. Mary Lyon suggested that LINE repeats may represent “way stations” along the X, which could signal or promote cis spreading (26). Autosomes are LINE poor when compared with the X, and it will be interesting to see whether the X short arm is also relatively LINE poor.
The large number of escapees on the human X chromosome has implications for understanding the pathogenesis of sex-chromosome disorders. As discussed by Carrel et al. (3), individuals with multiple copies of the X short arm are predicted—and indeed found—to have a more severe phenotype than individuals with multiple copies of the long arm. Increased expression of the many escapees on the X short arm would explain these different phenotypes. Further implications of Carrel et al.’s work can be inferred for the inheritance and phenotypes of X-linked diseases. In cases of genes subject to X inactivation, carriers of a disease allele often show skewing of X inactivation, resulting in a normal or near-normal phenotype. For some genes that escape, a dominant abnormal phenotype could be apparent in carrier females because of interference of an abnormal protein with the normal product. Carrier females with mutations in recessive genes that escape X inactivation would be protected by expression from the normal allele in all cells. Alternatively, if dosage is critical, females could be affected by haploinsufficiency of genes that escape. Such haploinsufficiency likely causes several Turner syndrome features, many of which appear to map to the X short arm.
Acknowledgments
P. A. Lingenfelter is gratefully acknowledged for her help in preparing the figures. This work was supported by National Institutes of Health Grant GM 46883.
Footnotes
See companion article on page 14440.
References
- 1.Lyon M. Nature (London) 1961;190:372–373. doi: 10.1038/190372a0. [DOI] [PubMed] [Google Scholar]
- 2.Disteche C M. Trends Genet. 1995;11:17–22. doi: 10.1016/s0168-9525(00)88981-7. [DOI] [PubMed] [Google Scholar]
- 3.Carrel L, Cottle A A, Goglin K C, Willard H F. Proc Natl Acad Sci USA. 1999;96:14440–14444. doi: 10.1073/pnas.96.25.14440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ohno S. Sex Chromosomes and Sex Linked Genes. Berlin: Springer; 1967. [Google Scholar]
- 5.Rice W R. BioScience. 1996;46:331–343. [Google Scholar]
- 6.Charlesworth B. Science. 1991;251:1030–1033. doi: 10.1126/science.1998119. [DOI] [PubMed] [Google Scholar]
- 7.Adler D A, Rugarli E I, Lingenfelter P A, Tsuchiya K, Poslinski D, Liggitt H D, Chapman V M, Elliott R W, Ballabio A, Disteche C M. Proc Natl Acad Sci USA. 1997;94:9244–9248. doi: 10.1073/pnas.94.17.9244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Graves J A. BioEssays. 1995;17:311–320. doi: 10.1002/bies.950170407. [DOI] [PubMed] [Google Scholar]
- 9.Lahn B T, Page D C. Science. 1999;286:877–879. doi: 10.1126/science.286.5441.964. [DOI] [PubMed] [Google Scholar]
- 10.Jegalian K, Page D C. Nature (London) 1998;394:776–780. doi: 10.1038/29522. [DOI] [PubMed] [Google Scholar]
- 11.Gartler S M, Dyer K A, Goldman M A. Mol Genet Med. 1992;2:121–160. doi: 10.1016/b978-0-12-462002-5.50010-8. [DOI] [PubMed] [Google Scholar]
- 12.Cooper D W, Johnston P G, Watson J M, Graves J A M. Semin Dev Biol. 1993;4:117–128. [Google Scholar]
- 13.White W M, Willard H F, Van Dyke D L, Wolff D J. Am J Hum Genet. 1998;63:20–28. doi: 10.1086/301922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huber R, Hansen R S, Strazzullo M, Pengue G, Mazzarella R, D’Urso M, Schlessinger D, Pilia G, Gartler S M, D’Esposito M. Pro Natl Acad Sci USA. 1999;96:616–621. doi: 10.1073/pnas.96.2.616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Salido E C, Yen P H, Koprivnikar K, Yu L C, Shapiro L J. Am J HumGenet. 1992;50:303–316. [PMC free article] [PubMed] [Google Scholar]
- 16.Watanabe M, Zinn A R, Page D C, Nishimoto T. Nat Genet. 1993;4:268–271. doi: 10.1038/ng0793-268. [DOI] [PubMed] [Google Scholar]
- 17.O’Brien S J, Menotti-Raymond M, Murphy W J, Nash W G, Wienberg J, Stanyon R, Copeland N G, Jenkins N A, Womack J E, Marshall Graves J A. Science. 1999;286:458–481. doi: 10.1126/science.286.5439.458. [DOI] [PubMed] [Google Scholar]
- 18.Disteche C M, Dinulos M B, Bassi M T, Elliott R W, Rugarli E I. Mamm Genome. 1998;9:1062–1064. doi: 10.1007/s003359900926. [DOI] [PubMed] [Google Scholar]
- 19.Zinn A R, Page D C, Fisher E M. Trends Genet. 1993;9:90–93. doi: 10.1016/0168-9525(93)90230-f. [DOI] [PubMed] [Google Scholar]
- 20.Duthie S M, Nesterova T B, Formstone E J, Keohane A M, Turner B M, Zakian S M, Brockdorff N. Hum Mol Genet. 1999;8:195–204. doi: 10.1093/hmg/8.2.195. [DOI] [PubMed] [Google Scholar]
- 21.Lee J T, Jaenisch R. Nature (London) 1997;386:275–279. doi: 10.1038/386275a0. [DOI] [PubMed] [Google Scholar]
- 22.Luoh S W, Jegalian K, Lee A, Chen E Y, Ridley A, Page D C. Genomics. 1995;29:353–363. doi: 10.1006/geno.1995.9994. [DOI] [PubMed] [Google Scholar]
- 23.Lingenfelter P A, Adler D A, Poslinski D, Thomas S, Elliott R W, Chapman V M, Disteche C M. Nat Genet. 1998;18:212–213. doi: 10.1038/ng0398-212. [DOI] [PubMed] [Google Scholar]
- 24.Brown C J, Carrel L, Willard H F. Am J Hum Genet. 1997;60:1333–1343. doi: 10.1086/515488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller A P, Willard H F. Proc Natl Acad Sci USA. 1998;95:8709–8714. doi: 10.1073/pnas.95.15.8709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lyon M F. Am J Hum Genet. 1998;63:17–19. doi: 10.1086/301940. [DOI] [PMC free article] [PubMed] [Google Scholar]