Abstract
As a major cardiac voltage-gated sodium channel isoform in the heart, Nav1.5 channel is essential for the cardiac action potential initiation and the subsequent propagation throughout the heart. Mutations of Nav1.5 have been linked to a variety of cardiac disease such as long QT syndrome (LQTs), Brugada syndrome, cardiac conduction defect, atrial fibrillation and dilated cardiomyopathy. Mutagenesis approach and heterologous expression systems are most frequently used to study the function of this channel. This review is primarily focused on recent findings on Nav1.5 mutations that are associated with type 3 long QT syndrome (LQT3) in particular. Understanding the functional changes of the Nav1.5 mutation may offer critical insight into the mechanism of long QT3 syndrome. In addition, this review will provide the updated information on the current progress of using various experimental model systems to study primarily the long QT3 syndrome.
Keywords: Cardiac sodium channel, cardiac arrhythmia, sodium channel mutation
Structure and physiological function of voltage-gated sodium channels
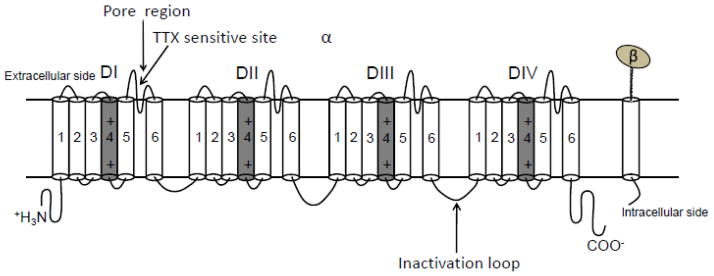
Voltage-gated sodium (Nav) channels initiate action potential depolarization and are responsible for propagating the action potential throughout the heart [43]. These channels are composed of a single pore forming α subunit with molecular weight 33–36 kD and may interacts with ancillary β subunits [7]. The α subunit consists of four structurally homologous transmembrane domains designated DI–DIV [46]. Each domain is composed of six putative transmembrane segments, which is named as S1–S6. The extracellular linker region between segment 5 and 6 within each domain joins together forming the channel pore referred to as “P loop” or “P segment”, which controls ion selectivity and permeation (Figure 1). Site-directed mutagenesis approach helped to identify the functional regions on S4 of the sodium channel [49]. Positively charged residues such as arginine or lysine at every third position (with mostly nonpolar residues intervening between these basic residues) on S4 segments are voltage sensors, which can lead to the fast activation of the sodium channel when the membrane potential is depolarized. The α subunit itself can form a functional channel, which is able to conduct ion and performs voltage-dependent gating process.
Figure 1.
Schematic linear representation of the structure of voltage-gated sodium channel. This figure is modified from the review article by Yu FH et al. [60]. The putative inactivation motif on the intracellular linker between DIII and DIV are indicated. The voltage sensor on segment 4 of each domain is marked with “+” and highlighted in grey. Extracellular loops between segments 5 and 6 within each domain forms the channel pore region. TTX sensitivity site is located in the extracellular loop of DI.
Upon activation, the channel is unable to open again due to the channel gating process called “inactivation”. This process varies from milliseconds (steady-state fast inactivation) to a sustained time up to a few seconds (slow inactivation). Inactivation is the process that causes the termination of the current flow [19]. Molecular biology and advanced biochemistry techniques allow us to know certain regions of the sodium channel that are responsible for channel fast inactivation. These regions are: the IFM motif with three hydrophobic amino acids Ile-Phe-Met [58] in the segment between transmembrane domain III and IV (DIII, DIV), the intracellular linker between S4 and S5 of DIII and DIV, the P loop, and the C-terminal domain of the channel [15].
As mentioned above, sodium channel also consists of auxiliary β subunits. So far, there are four sodium channel β subunits (β1 to β4) that have been identified. These four subunits can be divided into two groups. One group contains β1 (encoded by SCN1B, localized in brain neuronal tissue, skeletal muscle and cardiac tissue [31]), and β3 (encoded by SCN3B, localized primarily in neuronal tissue [36]; while the other group contains β2 and β4. β1 and β3 are very similar in amino acid sequence and they are noncovalently linked with α subunits [21, 36]. Studies indicates that the β subunit interacts with DI and DIV of the α subunit [32]. Co-expression of β1 subunit with α subunit modulates channel gating kinetics [32]. It has been shown that co-expression of the β1 subunit with the neuronal or skeletal muscle α subunit in Xenopus oocytes increases cell surface channel expression. Further, channel inactivation and activation rates were both altered. Channel inactivation curve was shifted to more negative potentials when β1 subunit was co-expressed with the cardiac sodium channel α subunit [31, 38]. Lori Isom’s group studied the expression of α and β subunits in heart tissue, and identified Nav1.1 α subunit, Nav1.5 α subunit, β1, and β2. Yet other groups have indentified the existence of Nav1.1, Nav1.3, Nav1.6 and Nav1.5 in cardiomyocytes [29, 30]. β3 has also been detected in the heart [12, 14, 36]. The developmental time course of β2 subunit expression suggests that it is at detectable levels from postnatal day 15. Both Nav1.5 and Nav1.1 were shown to be co-expressed with β1 and β2 subunits [12]. In cardiomyocytes, it was shown that, although the α subunit associates with both β1 and β2 subunits, only the β1 subunit has a modulatory effect on the electrophysiological properties of Nav1.5. The β2 subunit seems to have no detectable effects on electrophysiological properties of cardiac sodium channels, suggesting that effects of β2 in the heart in vivo may involve cell adhesion and cytoskeletal communication instead of modulating channel gating process [12].
So far, there are nine subtypes of voltage-gated sodium channels namely Nav1.1 to Nav1.9, and they are encoded by different genes [16]. Nav1.1, Nav1.2, Nav1.3, Nav1.6, Nav1.7, Nav1.8 and Nav1.9 are found in neurons. Nav1.1, Nav1.2 and Nav1.6 are found in the brain. Nav1.4 is found highly expressed in the skeletal muscle. In cardiac tissue, the predominant form of sodium channel is Nav1.5, which is encoded by SCN5A gene. Mutations of this gene have been linked to many types of cardiac arrhythmia such as long QT3 syndrome, atrial fibrillation, sick sinus node syndrome, Brugada syndrome, conduction defect, and dilated cardiomyopathy [5, 6, 11, 34, 47, 53, 54].
Cardiac sodium channel Nav1.5 tissue distribution
As mentioned above, the major cardiac sodium channel α subunit in the heart is Nav1.5 [60]. The α subunit is encoded by the SCN5A gene. Most mutations that affect the cardiac sodium channel are located on this gene. Cardiomyocytes also express neuronal Nav channel isoforms, such as Nav1.1, Nav1.3, and 1.6, but the location of these channels are mainly located on T-tubules [30]. The Nav1.5 isoform was found at high levels and is the major sodium channel isoform in adult rat heart [43]. Nav1.5 isoform is detectable in embryonic, neonatal skeletal muscle and denervated skeletal muscle [25]. However, it is not detectable in adult skeletal muscle. Like other types of voltage-gated sodium channels, these channels contain a principle α subunit and one or more auxiliary β subunits. The α subunit is the pore forming subunit, which can form a functional channel when it is expressed alone in the heterologous expression system. When expressed, this large α subunit accounts for the defining properties of voltage-gated sodium channels. It contains characteristic toxin-binding sites, channel pore, gates, and voltage sensors.
The tissue distribution of the cardiac sodium channel was demonstrated by both immunohistochemistry and conventional RT-PCR experiments. It was found that these channels are localized not only on the cardiomyocyte surface, but also on T-tubules. Later on, it was also demonstrated that cardiac sodium channels are concentrated at the terminal of the intercalated discs [9, 30], which separate adjacent cells and contain gap junctions that are responsible for propagating the cardiac action potential. The specific distribution of Nav1.5 as compared to the neuronal forms of sodium channel in the heart suggest that it may be crucial for action potential initiation while the action potential propagates from one cell to the other within the heart. However, the specialized distribution pattern of neuronal forms of sodium channels within the heart suggests that they are responsible for conducting action potentials inside of the myocyte to activate the contractile machinery [10].
Function of Nav1.5 on long QT3 syndromes
In the patient population that has been identified to carry LQTs, about 50% was associated with mutations on channels or associated proteins. Since the first identification of genetic mutations that cause LQTs in 1991, mutations have been discovered in 12 different genes. The classification of the inherited LQTs was based on the chronological order in which the location of the mutation was identified. The first SCN5A mutation was identified to be associated with LQT3 in 1995 by Wang et al [54]. Sodium channel mutations account for about 7% of the total identified LQTs-causing mutations. The typical ECG study for LQT3 indicates the long ST-segment interval before the onset of a late prominent T wave, which is the characteristic of LQT3 phenotype. Mutations in the alpha subunit of cardiac sodium channel have been identified and were linked to a series of other cardiac diseases such as Brugada syndrome, sick sinus node disease, cardiac conduction defect, dilated cardiomyopathy, and atrial fibrillation [40, 47, 48, 50].
LQT3 disease-causing gene was mapped to 3p21–24 by Mark Keating’s group using “candidate gene approach” [24]. Three amino acid KPQ deletions at position 1505–1507 of the α subunit in affected members of two distinct families with LQTs history were found by this group and this triple amino acid deletion is the most extensively characterized. This three amino acid in-frame deletion Lys-1505, Pro-1506, Gln-1507 was predicted to be in the cytoplasmic linker between DIII and DIV. The presence of exactly identical deletions in two unrelated families with LQT3 history provide strong evidence that SCN5A mutation is the most likely cause of LQT3. In 1995, Bennett et al. reported biophysical properties of this mutant channel. They showed that fast inactivation of the mutant channel is delayed since time constants for current decay were decreased in the ΔKPQ mutant (WT: τ fast = 1.47 ± 0.11 ms, τ slow = 8.59 ± 0.71 ms; ΔKPQ: τ fast = 0.98 ± 0.07 ms, τ slow = 5.40 ± 0.55 ms, p < 0.01; data are presented as means ± s.e.m.) not increased as was predicted. However, unlike WT channels, there are abnormal sustained sodium current (Isus) in ΔKPQ expressing cells. The Isus (~ 5% of the peak inward current at −20 mV) was shown not to decay within a 200 ms depolarization pulse [4, 52]. This sustained current, also called late INa was also found in a large number of other LQT3 related sodium channel mutations [2]. Single channel recordings indicated that ΔKPQ channels present multiple intermittent reopenings, which induces an Isus or delayed inactivation. Beside phenotypical sodium channel changes which are indicated by this first identified ΔKPQ sodium channel mutation, other gain-of-function electrophysiological changes including left-ward shift of the activation curve, increase of window current, faster recovery from inactivation and ramp current are also identified to be correlated with LQT3 syndrome [3, 20].
Voltage-gated sodium channels underline the rapid depolarization phase in ventricular cardiomyocytes and also conduct a small portion of current during the plateau phase of the action potential [1]. Therefore, a subtle abnormality of sodium channel function such as delayed sodium channel inactivation, or altered voltage dependence of channel inactivation could delay the cardiac repolarization phase. As a consequence, sodium channel functional defects can cause small increase in net inward sodium current and lead to QT interval prolongation and arrhythmia. Typically, the SCN5A mutations that are correlated with LQT3 are gain-of-function mutations (Table 1). Studies have demonstrated that gain-of-function mutations disturb the delicate balance between outward and inward currents which are involved in the plateau phase of the action potential repolarization process. In the presence of APD prolongation, there is a propensity for cardiomyocytes to develop early afterdepolarizations, which can trigger TdP. Under normal conditions, during the plateau phase nearly 99% of the channels become inactivated and transitioned into a non-conducting state while the remaining channels stay in the open activated state, sustaining a small window-current that contributes to the plateau phase. Therefore, mutations that disrupt the inactivation process will dramatically enhance the window-current and lead to the action potential prolongation [4].
Table 1.
List of representative SCN5A mutations that are associated with LQT3 syndrome.
| Mutation | Locus in Nav1.5 protein | Biophysical consequences | References |
|---|---|---|---|
| ΔKPQ1505–1507 | in the DIII–IV linker | ↑late INa, defect in inactivation, ↑rate of recovery from inactivation | Ref [8, 54] |
| F1473C | in the DIII–DIV linker | ↑late INa, depolarizing shift of the steady state inactivation, speeds the recovery from fast inactivation, uarr; ramp current | Ref [3] |
| F1473S | in the DIII–DIV linker | ↑late INa, depolarizing shift of the steady state inactivation | Ref [44] |
| ΔQKP1507–1509 | in the DIII–DIV linker | ↑late INa, depolarizing shift of the steady state activation | Ref [26] |
| I1768V | in DIVS6 near the C-terminal end | ↑speed of recovery from inactivation, and less slow inactivation | Ref [17] |
| V411M | in DIS6 | ↑late INa, depolarizing shift of the steady state activation | Ref [20] |
| L619F | in the DI-DII linker | ↑late INa, depolarizing shift of the steady state inactivation | Ref [56] |
Approaches that have been and are currently used to study mutation-induced LQTs including LQT3 syndrome
Heterologous expression system
Early studies on voltage-gated sodium channel were performed in vitro using Xenopus oocytes as a heterologous expression system [57, 58]. These studies offered detailed fundamental characterizations of the voltage-gated sodium channel. Human embryonic kidney (HEK) cells such as HEK293 cells are most frequently used to study functional changes following SCN5A mutations [3]. Other HEK cells such tSA cells and Chinese hamster ovary (CHO) cells are also used [33, 41] as heterologous expression systems. Together, these varieties of expression systems offer a platform to study the physiological function of the sodium channel and pathophysiological changes following gene mutations. However, the advantage of these systems is limited due to the fact that the cellular signaling component might be different from the cardiomyocyte.
Mouse model of LQT3
With the development of transgenic animal technique, experimental mouse models are emerging as a useful tool to study the LQT3 syndrome in vivo (Table 2). Transgenic mouse models carrying ΔKPQ and N1325S mutants were made [18, 39, 59]. In vivo studies from these mice indicated the presence of prolonged QT interval, spontaneous ventricular tachycardia and ventricular fibrillation. In vitro electrophysiology studies indicated that cardiomyocytes from these mice have prolonged APD, increased Isus, and early afterdepolarizations [13, 42, 51]. These animal models provide an opportunity to study the underlying mechanism of LQT3. However, limitation of the mouse model must be taken into account since there are significant electrophysiological differences between mouse hearts and human hearts [37]. Based on this, one must be very cautious while interpreting data generated using mouse model.
Table 2.
Representative mouse models of SCN5A related LQT3 syndrome.
| Model | Locus of the mutation | Genetic approach | Biophysical consequences | References |
|---|---|---|---|---|
| WT/Δ1505–1507KPQ | intracellular loop between DIII and DIV of Nav1.5 | knock-in | ↑late INa, | Ref [13, 39, 40] |
| WT/N1325S | intracellular region between S4–S5 of DIII of the Nav1.5 | transgenic | ↑late INa | Ref [51] |
| WT/1798insD | C-terminal domain | knock-in | ↑late INa, ↓INa | Ref [42] |
Computational approach
So far, a number of studies have used computational approach to simulate the action potential profile changes subsequent to Nav1.5 channel mutations and drug application [23, 55].The predicted computational analysis method allow to explore the consequences of disease-causing mutations on cellular electrical activity. One is beneficial not only from modeling the action potential profile itself but also from pacing the cell with different frequency and getting to know cellular activity changes as a consequence of gene mutation or drug application based on computational approach. Therefore, mechanistic insights on mutations can be obtained. Although this approach is widely used, yet ultimate changes of action potential profile are still hypothetical. Other approaches such as using primary myocytes are indeed needed to be explored.
Neonatal cardiomyocyte expression model
Recently, neonatal mouse cardiomyocytes have been used as a mammalian expression system to study ion channel mutations found in patients with LQTs [28]. In this study, fresh isolated cardiomyocytes from neonatal mouses were transiently transfected with the cDNA encoding the channel of interest using Amaxa Nucleotranfection kit (Lonza Inc.). The advantage of using neonatal cardiomyocytes as an expression system is that these cells have essential signaling components, which may play very crucial role in regulating ion channels. Although this approach has not been applied to study the sodium channel mutation that causes LQT3, this could be one of the future experimental approaches.
Recently, we have extended our study on SCN5A mutations using neonatal rat cardiomyocytes which express a mutant SCN5A cDNA. The mutation was previously identified in human patients. Ex vivo gene transfection method has been used by transfecting the gene using Nucleofection transfection kit. Using this approach, we have studied the functional consequence of a mutation on intracellular loop III and found that this mutation not only causes a series of gain-of-function changes in electrophysiological properties and gating process, but also leads to a significant prolongation of the action potential duration (unpublished data). Spontaneous early after depolarizations were also found (unpublished data). Although the exact copy number of the plasmids that were introduced into myocytes cannot be determined, this approach offers us a new strategy to study electrophysiological properties of the cardiac sodium channel using primary cells.
Future direction of modeling cardiac LQT3 syndrome with pluripotent stem cells
Itzhaki et al and Moretti et al established the most recent new paradigm for in vitro disease modeling of LQT1 and LQT2 syndromes [22, 35]. In their studies, induced pluoripotent stem cells (iPS cells) from patients suffering from the disease with characteristic of LQTs were differentiated into cardiomyocytes. This new advancement offers a new platform and strategy for mechanistically studying the channel mutation. In addition, it is useful for testing therapeutic compounds. In the future, iPS cell lines from the LQT3 patients can be used to differentiate into cardiomyocytes, and the pathophysiological changes that are caused by a specific mutation can be potentially explored. Therapeutic compounds can be tested on LQT3-based cardiomyocytes. Therefore, mutation-specific and individualized therapy can be potentially established.
Conclusions
This review summarizes the current understanding of sodium channel and focus particularly on their physiological and pathophysiological functions in cardiac arrhythmia. Various approaches to study the LQTs-related channel mutations have been discussed in this review. We have used a strategy to characterize the LQT3 syndrome disease-causing SCN5A mutations in neonatal rat cardiomyocytes. Functional changes of the action potential profile such as action potential duration prolongation and the onset of spontaneous early afterdepolarizations as a consequence of certain sodium channel mutations were identified. Therefore, our approach using neonatal rat cardiomyocytes to study the LQT3-related sodium channel mutations is a useful and effective strategy for analyzing the physiological phenotypes of various sodium channel mutations.
References
- 1.Attwell D, Cohen I, Eisner D, Ohba M, Ojeda C. The steady state TTX-sensitive (“window”) sodium current in cardiac Purkinje fibres. Pflugers Arch. 1979;379:137–142. doi: 10.1007/BF00586939. [DOI] [PubMed] [Google Scholar]
- 2.Bankston JR, Sampson KJ, Kateriya S, Glaaser IW, Malito DL, Chung WK, Kass RS. A novel LQT-3 mutation disrupts an inactivation gate complex with distinct rate-dependent phenotypic consequences. Channels (Austin) 2007;1:273–280. doi: 10.4161/chan.4956. [DOI] [PubMed] [Google Scholar]
- 3.Bankston JR, Yue M, Chung W, Spyres M, Pass RH, Silver E, Sampson KJ, Kass RS. A novel and lethal de novo LQT-3 mutation in a newborn with distinct molecular pharmacology and therapeutic response. PLoS One. 2007;2:e1258. doi: 10.1371/journal.pone.0001258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bennett PB, Yazawa K, Makita N, George AL., Jr Molecular mechanism for an inherited cardiac arrhythmia. Nature. 1995;376:683–685. doi: 10.1038/376683a0. [DOI] [PubMed] [Google Scholar]
- 5.Benson DW, Wang DW, Dyment M, Knilans TK, Fish FA, Strieper MJ, Rhodes TH, George AL., Jr Congenital sick sinus syndrome caused by recessive mutations in the cardiac sodium channel gene (SCN5A) J Clin Invest. 2003;112:1019–1028. doi: 10.1172/JCI18062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bezzina C, Veldkamp MW, van Den Berg MP, Postma AV, Rook MB, Viersma JW, van Langen IM, Tan-Sindhunata G, Bink-Boelkens MT, van Der Hout AH, Mannens MM, Wilde AA. A single Na(+) channel mutation causing both long-QT and Brugada syndromes. Circ Res. 1999;85:1206–1213. doi: 10.1161/01.res.85.12.1206. [DOI] [PubMed] [Google Scholar]
- 7.Catterall WA. From ionic currents to molecular mechanisms: the structure and function of voltage-gated sodium channels. Neuron. 2000;26:13–25. doi: 10.1016/s0896-6273(00)81133-2. [DOI] [PubMed] [Google Scholar]
- 8.Chandra R, Starmer CF, Grant AO. Multiple effects of KPQ deletion mutation on gating of human cardiac Na+ channels expressed in mammalian cells. Am J Physiol. 1998;274:H1643–1654. doi: 10.1152/ajpheart.1998.274.5.H1643. [DOI] [PubMed] [Google Scholar]
- 9.Cohen SA. Immunocytochemical localization of rH1 sodium channel in adult rat heart atria and ventricle. Presence in terminal intercalated disks. Circulation. 1996;94:3083–3086. doi: 10.1161/01.cir.94.12.3083. [DOI] [PubMed] [Google Scholar]
- 10.Cusdin FS, Clare JJ, Jackson AP. Trafficking and cellular distribution of voltage-gated sodium channels. Traffic. 2008;9:17–26. doi: 10.1111/j.1600-0854.2007.00673.x. [DOI] [PubMed] [Google Scholar]
- 11.Darbar D, Kannankeril PJ, Donahue BS, Kucera G, Stubblefield T, Haines JL, George AL, Jr, Roden DM. Cardiac sodium channel (SCN5A) variants associated with atrial fibrillation. Circulation. 2008;117:1927–1935. doi: 10.1161/CIRCULATIONAHA.107.757955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dhar Malhotra J, Chen C, Rivolta I, Abriel H, Malhotra R, Mattei LN, Brosius FC, Kass RS, Isom LL. Characterization of sodium channel alpha- and beta-subunits in rat and mouse cardiac myocytes. Circulation. 2001;103:1303–1310. doi: 10.1161/01.cir.103.9.1303. [DOI] [PubMed] [Google Scholar]
- 13.Fabritz L, Kirchhof P, Franz MR, Nuyens D, Rossenbacker T, Ottenhof A, Haverkamp W, Breithardt G, Carmeliet E, Carmeliet P. Effect of pacing and mexiletine on dispersion of repolarisation and arrhythmias in DeltaKPQ SCN5A (long QT3) mice. Cardiovasc Res. 2003;57:1085–1093. doi: 10.1016/s0008-6363(02)00839-8. [DOI] [PubMed] [Google Scholar]
- 14.Fahmi AI, Patel M, Stevens EB, Fowden AL, John JE, 3rd, Lee K, Pinnock R, Morgan K, Jackson AP, Vandenberg JI. The sodium channel beta-subunit SCN3b modulates the kinetics of SCN5a and is expressed heterogeneously in sheep heart. J Physiol. 2001;537:693–700. doi: 10.1111/j.1469-7793.2001.00693.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Glaaser IW, Bankston JR, Liu H, Tateyama M, Kass RS. A carboxyl-terminal hydrophobic interface is critical to sodium channel function. Relevance to inherited disorders. J Biol Chem. 2006;281:24015–24023. doi: 10.1074/jbc.M605473200. [DOI] [PubMed] [Google Scholar]
- 16.Goldin AL, Barchi RL, Caldwell JH, Hofmann F, Howe JR, Hunter JC, Kallen RG, Mandel G, Meisler MH, Netter YB, Noda M, Tamkun MM, Waxman SG, Wood JN, Catterall WA. Nomenclature of voltage-gated sodium channels. Neuron. 2000;28:365–368. doi: 10.1016/s0896-6273(00)00116-1. [DOI] [PubMed] [Google Scholar]
- 17.Groenewegen WA, Bezzina CR, van Tintelen JP, Hoorntje TM, Mannens MM, Wilde AA, Jongsma HJ, Rook MB. A novel LQT3 mutation implicates the human cardiac sodium channel domain IVS6 in inactivation kinetics. Cardiovasc Res. 2003;57:1072–1078. doi: 10.1016/s0008-6363(02)00838-6. [DOI] [PubMed] [Google Scholar]
- 18.Guzadhur L, Pearcey SM, Duehmke RM, Jeevaratnam K, Hohmann AF, Zhang Y, Grace AA, Lei M, Huang CL. Atrial arrhythmogenicity in aged Scn5a+/DeltaKPQ mice modeling long QT type 3 syndrome and its relationship to Na+ channel expression and cardiac conduction. Pflugers Arch. 2010;460:593–601. doi: 10.1007/s00424-010-0851-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hodgkin AL, Huxley AF. Currents carried by sodium and potassium ions through the membrane of the giant axon of Loligo. J Physiol. 1952;116:449–472. doi: 10.1113/jphysiol.1952.sp004717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Horne AJ, Eldstrom J, Sanatani S, Fedida D. A novel mechanism for LQT3 with 2:1 block: a pore-lining mutation in Nav1.5 significantly affects voltage-dependence of activation. Heart Rhythm. 2011;8:770–777. doi: 10.1016/j.hrthm.2010.12.041. [DOI] [PubMed] [Google Scholar]
- 21.Isom LL, De Jongh KS, Patton DE, Reber BF, Offord J, Charbonneau H, Walsh K, Goldin AL, Catterall WA. Primary structure and functional expression of the beta 1 subunit of the rat brain sodium channel. Science. 1992;256:839–842. doi: 10.1126/science.1375395. [DOI] [PubMed] [Google Scholar]
- 22.Itzhaki I, Maizels L, Huber I, Zwi-Dantsis L, Caspi O, Winterstern A, Feldman O, Gepstein A, Arbel G, Hammerman H, Boulos M, Gepstein L. Modelling the long QT syndrome with induced pluripotent stem cells. Nature. 2011;471:225–229. doi: 10.1038/nature09747. [DOI] [PubMed] [Google Scholar]
- 23.Jarecki BW, Piekarz AD, Jackson JO, 2nd, Cummins TR. Human voltage-gated sodium channel mutations that cause inherited neuronal and muscle channelopathies increase resurgent sodium currents. J Clin Invest. 2010;120:369–378. doi: 10.1172/JCI40801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang C, Atkinson D, Towbin JA, Splawski I, Lehmann MH, Li H, Timothy K, Taggart RT, Schwartz PJ, Vincent GM, et al. Two long QT syndrome loci map to chromosomes 3 and 7 with evidence for further heterogeneity. Nat Genet. 1994;8:141–147. doi: 10.1038/ng1094-141. [DOI] [PubMed] [Google Scholar]
- 25.Kallen RG, Sheng ZH, Yang J, Chen LQ, Rogart RB, Barchi RL. Primary structure and expression of a sodium channel characteristic of denervated and immature rat skeletal muscle. Neuron. 1990;4:233–242. doi: 10.1016/0896-6273(90)90098-z. [DOI] [PubMed] [Google Scholar]
- 26.Keller DI, Acharfi S, Delacretaz E, Benammar N, Rotter M, Pfammatter JP, Fressart V, Guicheney P, Chahine M. A novel mutation in SCN5A, delQKP 1507–1509, causing long QT syndrome: role of Q1507 residue in sodium channel inactivation. J Mol Cell Cardiol. 2003;35:1513–1521. doi: 10.1016/j.yjmcc.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 27.Leffler A, Herzog RI, Dib-Hajj SD, Waxman SG, Cummins TR. Pharmacological properties of neuronal TTX-resistant sodium channels and the role of a critical serine pore residue. Pflugers Arch. 2005;451:454–463. doi: 10.1007/s00424-005-1463-x. [DOI] [PubMed] [Google Scholar]
- 28.Lin EC, Holzem KM, Anson BD, Moungey BM, Balijepalli SY, Tester DJ, Ackerman MJ, Delisle BP, Balijepalli RC, January CT. Properties of WT and mutant hERG K(+) channels expressed in neonatal mouse cardiomyocytes. Am J Physiol Heart Circ Physiol. 2010;298:H1842–1849. doi: 10.1152/ajpheart.01236.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maier SK, Westenbroek RE, Schenkman KA, Feigl EO, Scheuer T, Catterall WA. An unexpected role for brain-type sodium channels in coupling of cell surface depolarization to contraction in the heart. Proc Natl Acad Sci U S A. 2002;99:4073–4078. doi: 10.1073/pnas.261705699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maier SK, Westenbroek RE, McCormick KA, Curtis R, Scheuer T, Catterall WA. Distinct subcellular localization of different sodium channel alpha and beta subunits in single ventricular myocytes from mouse heart. Circulation. 2004;109:1421–1427. doi: 10.1161/01.CIR.0000121421.61896.24. [DOI] [PubMed] [Google Scholar]
- 31.Makita N, Bennett PB, Jr, George AL., Jr Voltage-gated Na+ channel beta 1 subunit mRNA expressed in adult human skeletal muscle, heart, and brain is encoded by a single gene. J Biol Chem. 1994;269:7571–7578. [PubMed] [Google Scholar]
- 32.Makita N, Bennett PB, George AL., Jr Molecular determinants of beta 1 subunit-induced gating modulation in voltage-dependent Na+ channels. J Neurosci. 1996;16:7117–7127. doi: 10.1523/JNEUROSCI.16-22-07117.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Makita N, Behr E, Shimizu W, Horie M, Sunami A, Crotti L, Schulze-Bahr E, Fukuhara S, Mochizuki N, Makiyama T, Itoh H, Christiansen M, McKeown P, Miyamoto K, Kamakura S, Tsutsui H, Schwartz PJ, George AL, Jr, Roden DM. The E1784K mutation in SCN5A is associated with mixed clinical phenotype of type 3 long QT syndrome. J Clin Invest. 2008;118:2219–2229. doi: 10.1172/JCI34057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McNair WP, Sinagra G, Taylor MR, Di Lenarda A, Ferguson DA, Salcedo EE, Slavov D, Zhu X, Caldwell JH, Mestroni L. SCN5A mutations associate with arrhythmic dilated cardiomyopathy and commonly localize to the voltage-sensing mechanism. J Am Coll Cardiol. 2011;57:2160–2168. doi: 10.1016/j.jacc.2010.09.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moretti A, Bellin M, Welling A, Jung CB, Lam JT, Bott-Flugel L, Dorn T, Goedel A, Hohnke C, Hofmann F, Seyfarth M, Sinnecker D, Schomig A, Laugwitz KL. Patient-specific induced pluripotent stem-cell models for long-QT syndrome. N Engl J Med. 2010;363:1397–1409. doi: 10.1056/NEJMoa0908679. [DOI] [PubMed] [Google Scholar]
- 36.Morgan K, Stevens EB, Shah B, Cox PJ, Dixon AK, Lee K, Pinnock RD, Hughes J, Richardson PJ, Mizuguchi K, Jackson AP. beta 3: an additional auxiliary subunit of the voltage-sensitive sodium channel that modulates channel gating with distinct kinetics. Proc Natl Acad Sci U S A. 2000;97:2308–2313. doi: 10.1073/pnas.030362197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nerbonne JM. Studying cardiac arrhythmias in the mouse--a reasonable model for probing mechanisms? Trends Cardiovasc Med. 2004;14:83–93. doi: 10.1016/j.tcm.2003.12.006. [DOI] [PubMed] [Google Scholar]
- 38.Nuss HB, Chiamvimonvat N, Perez-Garcia MT, Tomaselli GF, Marban E. Functional association of the beta 1 subunit with human cardiac (hH1) and rat skeletal muscle (mu 1) sodium channel alpha subunits expressed in Xenopus oocytes. J Gen Physiol. 1995;106:1171–1191. doi: 10.1085/jgp.106.6.1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nuyens D, Stengl M, Dugarmaa S, Rossenbacker T, Compernolle V, Rudy Y, Smits JF, Flameng W, Clancy CE, Moons L, Vos MA, Dewerchin M, Benndorf K, Collen D, Carmeliet E, Carmeliet P. Abrupt rate accelerations or premature beats cause life-threatening arrhythmias in mice with long-QT3 syndrome. Nat Med. 2001;7:1021–1027. doi: 10.1038/nm0901-1021. [DOI] [PubMed] [Google Scholar]
- 40.Papadatos GA, Wallerstein PM, Head CE, Ratcliff R, Brady PA, Benndorf K, Saumarez RC, Trezise AE, Huang CL, Vandenberg JI, Colledge WH, Grace AA. Slowed conduction and ventricular tachycardia after targeted disruption of the cardiac sodium channel gene Scn5a. Proc Natl Acad Sci U S A. 2002;99:6210–6215. doi: 10.1073/pnas.082121299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Persson F, Andersson B, Duker G, Jacobson I, Carlsson L. Functional effects of the late sodium current inhibition by AZD7009 and lidocaine in rabbit isolated atrial and ventricular tissue and Purkinje fibre. Eur J Pharmacol. 2007;558:133–143. doi: 10.1016/j.ejphar.2006.11.040. [DOI] [PubMed] [Google Scholar]
- 42.Remme CA, Verkerk AO, Nuyens D, van Ginneken AC, van Brunschot S, Belterman CN, Wilders R, van Roon MA, Tan HL, Wilde AA, Carmeliet P, de Bakker JM, Veldkamp MW, Bezzina CR. Overlap syndrome of cardiac sodium channel disease in mice carrying the equivalent mutation of human SCN5A-1795insD. Circulation. 2006;114:2584–2594. doi: 10.1161/CIRCULATIONAHA.106.653949. [DOI] [PubMed] [Google Scholar]
- 43.Rogart RB, Cribbs LL, Muglia LK, Kephart DD, Kaiser MW. Molecular cloning of a putative tetrodotoxin-resistant rat heart Na+ channel isoform. Proc Natl Acad Sci U S A. 1989;86:8170–8174. doi: 10.1073/pnas.86.20.8170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ruan Y, Denegri M, Liu N, Bachetti T, Seregni M, Morotti S, Severi S, Napolitano C, Priori SG. Trafficking defects and gating abnormalities of a novel SCN5A mutation question gene-specific therapy in long QT syndrome type 3. Circ Res. 2010;106:1374–1383. doi: 10.1161/CIRCRESAHA.110.218891. [DOI] [PubMed] [Google Scholar]
- 45.Satin J, Kyle JW, Chen M, Bell P, Cribbs LL, Fozzard HA, Rogart RB. A mutant of TTX-resistant cardiac sodium channels with TTX-sensitive properties. Science. 1992;256:1202–1205. doi: 10.1126/science.256.5060.1202. [DOI] [PubMed] [Google Scholar]
- 46.Sato C, Sato M, Iwasaki A, Doi T, Engel A. The sodium channel has four domains surrounding a central pore. J Struct Biol. 1998;121:314–325. doi: 10.1006/jsbi.1998.3990. [DOI] [PubMed] [Google Scholar]
- 47.Schott JJ, Alshinawi C, Kyndt F, Probst V, Hoorntje TM, Hulsbeek M, Wilde AA, Escande D, Mannens MM, Le Marec H. Cardiac conduction defects associate with mutations in SCN5A. Nat Genet. 1999;23:20–21. doi: 10.1038/12618. [DOI] [PubMed] [Google Scholar]
- 48.Smits JP, Koopmann TT, Wilders R, Veldkamp MW, Opthof T, Bhuiyan ZA, Mannens MM, Balser JR, Tan HL, Bezzina CR, Wilde AA. A mutation in the human cardiac sodium channel (E161K) contributes to sick sinus syndrome, conduction disease and Brugada syndrome in two families. J Mol Cell Cardiol. 2005;38:969–981. doi: 10.1016/j.yjmcc.2005.02.024. [DOI] [PubMed] [Google Scholar]
- 49.Stuhmer W, Conti F, Suzuki H, Wang XD, Noda M, Yahagi N, Kubo H, Numa S. Structural parts involved in activation and inactivation of the sodium channel. Nature. 1989;339:597–603. doi: 10.1038/339597a0. [DOI] [PubMed] [Google Scholar]
- 50.Tan HL, Bink-Boelkens MT, Bezzina CR, Viswanathan PC, Beaufort-Krol GC, van Tintelen PJ, van den Berg MP, Wilde AA, Balser JR. A sodium-channel mutation causes isolated cardiac conduction disease. Nature. 2001;409:1043–1047. doi: 10.1038/35059090. [DOI] [PubMed] [Google Scholar]
- 51.Tian XL, Yong SL, Wan X, Wu L, Chung MK, Tchou PJ, Rosenbaum DS, Van Wagoner DR, Kirsch GE, Wang Q. Mechanisms by which SCN5A mutation N1325S causes cardiac arrhythmias and sudden death in vivo. Cardiovasc Res. 2004;61:256–267. doi: 10.1016/j.cardiores.2003.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang DW, Yazawa K, George AL, Jr, Bennett PB. Characterization of human cardiac Na+ channel mutations in the congenital long QT syndrome. Proc Natl Acad Sci U S A. 1996;93:13200–13205. doi: 10.1073/pnas.93.23.13200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang DW, Makita N, Kitabatake A, Balser JR, George AL., Jr Enhanced Na(+) channel intermediate inactivation in Brugada syndrome. Circ Res. 2000;87:E37–43. doi: 10.1161/01.res.87.8.e37. [DOI] [PubMed] [Google Scholar]
- 54.Wang Q, Shen J, Splawski I, Atkinson D, Li Z, Robinson JL, Moss AJ, Towbin JA, Keating MT. SCN5A mutations associated with an inherited cardiac arrhythmia, long QT syndrome. Cell. 1995;80:805–811. doi: 10.1016/0092-8674(95)90359-3. [DOI] [PubMed] [Google Scholar]
- 55.Wehrens XH, Abriel H, Cabo C, Benhorin J, Kass RS. Arrhythmogenic mechanism of an LQT-3 mutation of the human heart Na(+) channel alpha-subunit: A computational analysis. Circulation. 2000;102:584–590. doi: 10.1161/01.cir.102.5.584. [DOI] [PubMed] [Google Scholar]
- 56.Wehrens XH, Rossenbacker T, Jongbloed RJ, Gewillig M, Heidbuchel H, Doevendans PA, Vos MA, Wellens HJ, Kass RS. A novel mutation L619F in the cardiac Na+ channel SCN5A associated with long-QT syndrome (LQT3): a role for the I-II linker in inactivation gating. Hum Mutat. 2003;21:552. doi: 10.1002/humu.9136. [DOI] [PubMed] [Google Scholar]
- 57.Wei J, Wang DW, Alings M, Fish F, Wathen M, Roden DM, George AL., Jr Congenital long-QT syndrome caused by a novel mutation in a conserved acidic domain of the cardiac Na+ channel. Circulation. 1999;99:3165–3171. doi: 10.1161/01.cir.99.24.3165. [DOI] [PubMed] [Google Scholar]
- 58.West JW, Patton DE, Scheuer T, Wang Y, Goldin AL, Catterall WA. A cluster of hydrophobic amino acid residues required for fast Na(+)-channel inactivation. Proc Natl Acad Sci U S A. 1992;89:10910–10914. doi: 10.1073/pnas.89.22.10910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yong SL, Ni Y, Zhang T, Tester DJ, Ackerman MJ, Wang QK. Characterization of the cardiac sodium channel SCN5A mutation, N1325S, in single murine ventricular myocytes. Biochem Biophys Res Commun. 2007;352:378–383. doi: 10.1016/j.bbrc.2006.11.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yu FH, Catterall WA. Overview of the voltage-gated sodium channel family. Genome Biol. 2003;4:207. doi: 10.1186/gb-2003-4-3-207. [DOI] [PMC free article] [PubMed] [Google Scholar]