Abstract
Objectives
To investigate whether (1) machine learning classifiers can help identify nonrandomized studies eligible for full-text screening by systematic reviewers; (2) classifier performance varies with optimization; and (3) the number of citations to screen can be reduced.
Methods
We used an open-source, data-mining suite to process and classify biomedical citations that point to mostly nonrandomized studies from 2 systematic reviews. We built training and test sets for citation portions and compared classifier performance by considering the value of indexing, various feature sets, and optimization. We conducted our experiments in 2 phases. The design of phase I with no optimization was: 4 classifiers × 3 feature sets × 3 citation portions. Classifiers included k-nearest neighbor, naïve Bayes, complement naïve Bayes, and evolutionary support vector machine. Feature sets included bag of words, and 2- and 3-term n-grams. Citation portions included titles, titles and abstracts, and full citations with metadata. Phase II with optimization involved a subset of the classifiers, as well as features extracted from full citations, and full citations with overweighted titles. We optimized features and classifier parameters by manually setting information gain thresholds outside of a process for iterative grid optimization with 10-fold cross-validations. We independently tested models on data reserved for that purpose and statistically compared classifier performance on 2 types of feature sets. We estimated the number of citations needed to screen by reviewers during a second pass through a reduced set of citations.
Results
In phase I, the evolutionary support vector machine returned the best recall for bag of words extracted from full citations; the best classifier with respect to overall performance was k-nearest neighbor. No classifier attained good enough recall for this task without optimization. In phase II, we boosted performance with optimization for evolutionary support vector machine and complement naïve Bayes classifiers. Generalization performance was better for the latter in the independent tests. For evolutionary support vector machine and complement naïve Bayes classifiers, the initial retrieval set was reduced by 46% and 35%, respectively.
Conclusions
Machine learning classifiers can help identify nonrandomized studies eligible for full-text screening by systematic reviewers. Optimization can markedly improve performance of classifiers. However, generalizability varies with the classifier. The number of citations to screen during a second independent pass through the citations can be substantially reduced.
Keywords: medical informatics, clinical research informatics, text mining, document classification, systematic reviews
1. Introduction
Translation of biomedical research into practice depends in part on the production of systematic reviews that synthesize available evidence for clinicians, researchers, and policymakers. Unfortunately, remarkable growth in the number of reviews has not kept pace with growth in the number of medical trials, which are sources of evidence [1]. The problem is even more serious because most reviews are traditional rather than systematic. What is needed is streamlined production of the latter [1, 2] to better control known threats to validity [3] while promoting transparent and reproducible science.
To support the creation and maintenance of quality systematic reviews (also known as evidence reports or comparative effectiveness reviews), a global network of Cochrane entities [4] and a North American network of AHRQ-funded Evidence-based Practice Centers [5, 6] exist. Even so, production is slow. For example, Tricco et al [7] report that 19% of protocols published in the respected Cochrane Library fail to reach fruition as full reviews. Of those that are published as reviews, the average time to completion is 2.4 years with a reported maximum of 9 years, which is the ceiling imposed by the study design. Worse, these estimates ignore time spent exploring the literature to assess significance of possible review questions, and then time spent developing a protocol.
A major bottleneck occurs when teammates screen studies. In a two-step process involving independent and replicated effort, teammates first identify provisionally eligible studies by reading typically thousands of citations. Then they repeat the process by reading full texts of studies identified in the first step to select the final set of studies for inclusion in a review. In other words, to be included in a review, a study must first appear to meet eligibility criteria based on reading its citation; if so, it is eligible for full-text review and provisionally eligible for inclusion in the systematic review. However, not until the full text of its report has been carefully considered in light of the protocol is a final decision made whether to include a study.
In a best-case scenario, teammates compare their decisions and resolve their differences after each step, usually by discussion. It is worth noting that screening procedures vary. For example, some review teams will consider a study for full-text review if at least one teammate thinks the citation (title plus abstract) appears to meet eligibility criteria. In contrast, other teams work to reach consensus when screening citations before they will consider a study worth reading as full text. Presumably, the latter procedure for screening citations is more labor intensive. The point is that workflow patterns vary by review team and topic (A. McKibbon, PhD, written communication, December 2010). Furthermore, it is likely that estimates of workload for professional review teams associated with established centers are underestimates for inexperienced volunteer teams that may be conducting one-off reviews, e.g., when launching new research programs.
The research that serves as the foundation for this study was conducted by Aphinyanaphongs et al [8], and later extended by Kilicoglu et al [9]. Their work entailed supervised machine learning methods and natural language processing to identify rigorous clinical trials in broad domains, such as therapy, rather than topical domains defined by review questions. Based on the work of Haynes and colleagues in a series of papers (e.g., see [10]), rigor was presumed if trials comparing treatments were randomized and controlled. However, identifying nonrandomized (NR) studies for inclusion in systematic reviews is an important problem because randomized controlled trials (RCTs) may be unlikely or even unethical for some research questions [11, 12]. For example, NR studies, such as case-control, cross-sectional, and cohort studies, are commonly employed to investigate exposure to environmental hazards, diagnostic test accuracy, disease etiology, human development, invasive surgery, adverse events, and rare disorders. Notably, in what is perhaps the first study to use machine learning methods to identify topically relevant trials for inclusion in systematic reviews, classification involved randomized and controlled drug trials [13], which is in keeping with the foundational research of [8].
For many review questions, the classification task involves a mix of designs because reviewers search for NR studies (if eligible) in addition to RCTs. The latter are preferred because they tend to be less biased relative to NR studies. However, when NR studies are eligible for inclusion in a systematic review, the Cochrane Non-Randomised Studies Methods Group enjoins investigators to not include design terms in their search filters [12]. Although filters exist to reliably retrieve RCTs [14], filters “to identify other study types are limited” (Appendix 2 in [15]; see also [11]). This is true even though development of filters is ongoing (e.g., see [16–20]). Thus, the initial screening phase can be more labor intensive when NR studies are eligible. In response to this dilemma, some of the Cochrane Review Groups allow NR design terms when the retrieval set is so large that the review becomes impractical (e.g., see [21]). If we take seriously the preference for not including design terms in searches for NR studies, an informatics solution to assist review teams seems especially warranted.
Researchers interested in [semi-]automating the screening phase for systematic reviews are currently using the classifiers complement naïve Bayes (cNB) [22] or a Support Vector Machine (SVM) with a linear kernel [23, 24], or are developing a factorized version of cNB [25]. The fact that these researchers are using different classifiers for their specific tasks indicates that understanding relative classifier performance is a necessary step for our task. Thus, we are interested in empirically comparing the performance of several supervised machine learning classifiers for a binary classification task using biomedical citations from extant systematic reviews. The task is binary because we want to classify primary studies as being eligible or not for further consideration by the review team. We also consider no optimization vs. optimization of features and parameters. Interestingly, a comparative study of classifiers by Colas and Brazdil [26] is sometimes cited as support for using a particular classifier. They found that an optimized k-nearest neighbor (k-NN) or naïve Bayes (NB) classifier could be as good as a linear SVM based on 20,000 newsgroup e-mails. However, they cautioned that their results should be validated for other document classification tasks to ensure generalizability. In sum, classifiers useful for newsgroup e-mail may not be as useful for biomedical citations. Thus, comparative studies of classifiers are warranted.
In general, our motivation for conducting this research is similar to that of other groups [13, 22–25], i.e., we want to facilitate production of systematic reviews. However, we are interested in assisting reviewers (regardless of experience or affiliation) by identifying classifiers that can reduce the number of citations that must be screened during a second independent pass through a set of citations. We interpret the usefulness of a classifier with respect to reducing the number of citations to screen rather than time spent screening because of differences in procedures, reviewer expertise, and number of teammates available for dividing the labor. In other words, valid baseline estimates of time spent screening and subsequent reductions in time depend on several variables that are not the focus of this study.
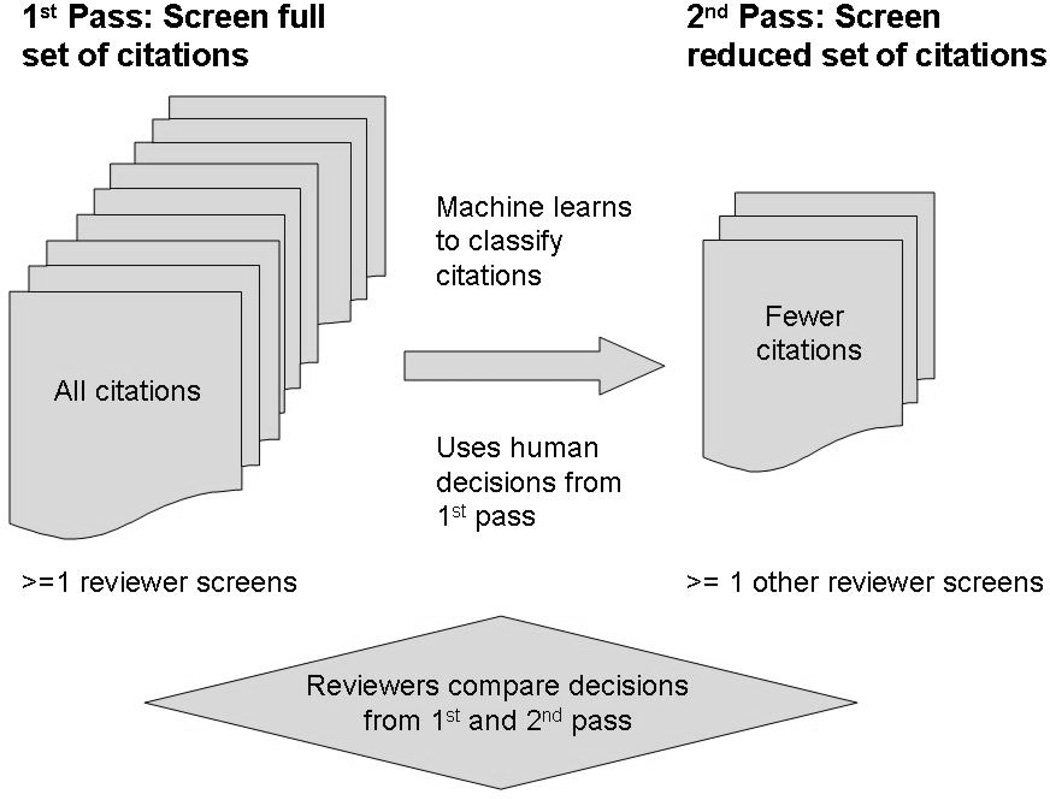
Additionally, until this relatively new area of translational informatics research matures, we assume that reviewers will insist on at least one complete cycle where human(s) screen the full set of citations. We further assume that a team consists of at least two people to ensure independent and replicated screening. In reality, more than one teammate can screen citations for the first pass as long as other people independently screen the same citations during the second pass. This procedure is meant to control random errors and bias introduced by humans. However, there are times when even two people cannot independently screen the entire set of citations. When this is the case, Cochrane suggests “a second person look at a sample [emphasis added] of the records” [27]. This is precisely our intention, i.e., we envision a machine learning system that returns a reduced set or sample of citations to screen for the second pass. The reduced set would include most if not all of the citations labeled as eligible for full-text review, as well as a subset of those labeled as ineligible during the initial screening. Human reviewers would still have to reach consensus regarding discrepant eligibility decisions from the first pass through the entire set when compared to a second pass through the reduced set (see Figure 1). Assisting reviewers in this way would enable a more focused, independent screening of citations during the second pass. Reviewer bias and error would be controlled, in part, because of the opportunity for a second screening by different teammate(s) who could potentially identify studies overlooked by the first reviewers. Furthermore, the workload would be reduced because the disproportionately large set of citations identified as ineligible by both humans and machine would be eliminated from further consideration.
Fig. 1.
Machine learning can reduce the number of citations needed to screen by systematic reviewers.
In sum, we conducted this study to investigate whether (1) machine learning classifiers can help identify NR studies eligible for full-text screening by systematic reviewers; (2) classifier performance varies with optimization of parameters and features extracted from biomedical citations; and (3) the number of citations to screen can be reduced. We did this by empirically comparing classifier performance using citations that point to mostly NR studies, varying optimization conditions, and then estimating the reduction in the number of citations to screen for the best classifier.
2. Methods
The citations for this study were from 2 Cochrane systematic reviews. One has to do with surgical interventions for treating ameloblastomas of the jaws [21] and the other with vaccines for preventing influenza in the elderly [28]. By using citations from extant systematic reviews, we capitalized on domain-specific knowledge. This is because citations were initially retrieved by Cochrane trials search coordinators who developed filters given reviewers’ knowledge of their topics.
For the ameloblastoma dataset, we had access to the entire set of citations (N=1815) retrieved from MEDLINE [29], EMBASE [30], the Cochrane Central Register of Controlled Trials, and the Cochrane Oral Health Group Trials Register. For the influenza dataset, we retrieved 5485 citations (94%) by re-running published MEDLINE and EMBASE searches. We also manually searched for 147 studies not in our retrieval set, but listed in the review as eligible for further consideration.
We managed citations in EndNote and recorded decisions as either exclude or include. Decisions were based on the consensus of at least 2 reviewers in the published author lists regarding eligibility [21, 28, 31]. From EndNote, we exported each corpus as a text file in MEDLINE format. We then created 3 text files for each citation: (1) the full citation, including title, abstract, and metadata (FULL); (2) the title and abstract (TIABS); and (3) the title (TITLE). We built training and test sets for each type of text file by randomly assigning files using a 2:1 split, respectively. To ensure comparability across training and test sets, we used the same random assignment for citation portions.
For the ameloblastoma review, the training set for each citation portion consisted of 1209 files: exclude=1133; include=76 (6.3%). The test set for each portion consisted of 606 files: exclude=567; include=39 (6.4%). For the influenza review, the training set consisted of 3679 files: exclude=3469; include=210 (5.7%); the test set consisted of 1806 files: exclude=1699; include=107 (5.9%). The citations labeled as include point to studies eligible for full text-review, as well as being provisionally eligible for eventual inclusion in the systematic review.
To extract features (processed words) and classify studies, we used the open-source, data-mining suite RapidMiner v.4.6 [32, 33] with a text plugin [34]. We processed text to create weighted feature vectors that represent each citation portion. This involved tokenizing (splitting up) strings of text, converting to lower case, filtering out stopwords and tokens with length less than 3, Porter stemming, and pruning out tokens that occurred in at most 3 citations. Features were weighted with TFIDF weights ([35], p.109), which are the product of term frequencies (TF) and inverse document frequencies (IDF). Note that for citations retrieved from MEDLINE or EMBASE, the metadata include tags and indexing terms from MeSH [36] or EMTREE [37], respectively. For this study, we treated metadata as any other text without preserving the tags, such as the MeSH tags TI for title or SO for source.
In general, we first trained a set of classifiers known to work well with text [26, 38] using processed features extracted from citations. Then we independently tested classifier models on a third of the data reserved for this purpose. We compared performance with respect to recall, precision, and a summary measure that overweights recall relative to precision. We chose to overweight recall because this is in keeping with the human goal of near-perfect recall when screening citations [12, 25, 39]. Human reviewers are overly inclusive during this phase in order to reduce the risk of overlooking relevant studies. This means that precision is sacrificed for recall. During their full-text review of studies identified by screening citations, reviewers effectively improve precision by eliminating studies that do not meet their inclusion criteria. Thus, for our purposes, we wanted to find classifiers with nearly perfect recall and precision good enough to reduce the number of citations to screen.
We conducted this study in two phases. Phase I involved neither optimization nor validation; phase II involved optimization of features and classifier parameters with cross-validation. In both phases, we conducted independent tests on the reserved data.
We defined best models as returning highest recall with precision good enough to reduce workload. Specifically, recall had to be at least 95% and precision had to be greater than 7% and 6% for the ameloblastoma and influenza datasets, respectively. The rationale for the cutoffs is as follows: when a model returns nearly perfect recall but poor precision, almost all of the eligible studies are identified along with many falsely identified ones. In the extreme, if precision equals the percentage eligible, the returned set of studies is as large as the entire set and no reduction in workload is possible. Thus, precision must surpass the percentage of studies identified by humans as being eligible for full-text review. Note that when recall is 95%, the machine falsely excludes 5% of the eligible studies. However, in comparing discrepant decisions, human(s) would reconsider the 5% they had identified but the machine had missed.
In our experiments, we compared the following classifiers: k-NN [26], NB [40], cNB [41], and evolutionary support vector machine (EvoSVM) [42]. NB and cNB are probabilistic learners; EvoSVM is functional; and k-NN is a lazy learner that classifies based on similarity or distance measures. Further, NB assumes conditional and positional independence of features; thus, the immediate context of features or processed words extracted from citations is ignored. cNB is suitable for imbalanced data and presumably more appropriate for this task because the percentage of eligible studies in systematic reviews is usually relatively small. Additionally, cNB relaxes the particularly unrealistic assumptions of NB regarding independence of features extracted from text written by humans. EvoSVM uses a kernel function to find a nonlinear hyperplane that maximally separates classes of documents. EvoSVM generalizes support vector machine classifiers and can optimize non-positive semi-definite kernel functions [42].
We used RapidMiner default settings for classifier parameters with the following exceptions: For EvoSVM, we set C=1 instead of C=0 in phase I based on [42]. In phase II, we set C=1, 10, or 20. The parameter C is a regularization constant that sets an upper bound for multipliers used in maximizing the margin between classes (cf. chapter 15 in [35]). For k-NN, we used cosine similarity measures instead of mixed Euclidean distances.
For both phases, performance measures included recall, precision, and an overall performance measure (F3), which is a weighted harmonic mean ([35], p.144). The formula for F is:
| (1) |
where beta is a non-negative number. Note that the notation F1 or F3 is short for Fbeta=1 or Fbeta=3, respectively. Thus, the formula for F3 is:
| (2) |
We estimated F3 rather than the traditional measure F1 that equally weights recall and precision. Although the relative weighting is more obvious when beta is expressed in terms of alpha (cf. [35], p. 144), the formulas presented are more common. In our opinion, F1 is inappropriate for this task because it is not in keeping with reviewer behavior during the screening phase.
2.1 Design of phase I (no optimization)
The design of phase I was: 4 classifiers × 3 citation portions × 3 feature sets. We used the ameloblastoma citations and did not optimize features or classifier parameters.
Classifiers included k-NN, NB, cNB, and EvoSVM. In early analyses, LibSVM with a radial or polynomial kernel either failed or returned very poor performance. We therefore dropped LibSVM from subsequent analyses.
Citation portions included TITLES, TIABS, and FULL citations.
Feature sets included unigrams or bag of single words (BOW), and 2-term (2G) and 3-term n-grams (3G). N-gram sets are hierarchical and therefore consist of features from previous set(s). For example, a 3G set consists of contiguous triples and pairs, as well as single processed features, i.e., trigrams, bigrams, and unigrams. We had competing reasons for comparing these feature sets. On the one hand, 2G or 3G could add linguistic phrases that improve classification; on the other, BOW could reduce computational burden.
Varying feature sets and citation portions allowed comparison of their relative contribution to classification. We expected that an n-gram feature set extracted from FULL citations would improve classifier performance. We reasoned that 2G or 3G sets would preserve some of the information in the indexing terms or phrases found in the metadata of FULL citations and, therefore, this feature–citation portion combination would be associated with better performance.
2.2 Design of phase II (optimization with cross-validation)
In this phase, we used ameloblastoma and influenza citations. We considered 3 classifiers, 2 feature sets, and 1 citation portion. Classifiers included k-NN, cNB, and EvoSVM. Given the results from phase I, we dropped NB and used BOW extracted from FULL citations. We also developed a second feature set by adding 2G title features to the BOW. This enrichment overweighted titles and added contextual information residing in pairs of title words.
For each information gain (IG) threshold, we selected features if the absolute value of the IG weight was >= to the threshold. We manually set the IG threshold outside of a loop for grid optimization of classifier parameters with an inner loop for 10-fold cross-validations.
We used the RapidMiner operator Grid Parameter Optimization to find the best parameter set per information threshold. This operator searches over a grid of parameter combinations to return an optimal set. Given the nature of human screening behavior, we searched for optimal sets yielding highest recall with precision greater than the cutoff. The size of the grid is determined by the levels of the parameters under consideration. For example, if one combines 2 parameters with 3 possible values each, the search is over a 3 × 3 grid with 9 cells. For each cell, an n-fold cross-validation is run. In our experiments, the total number of runs (N) for each classifier equals the number of IG thresholds × the number of cells in the grid × the number of folds in the cross-validations. For example, N=480 (k-NN), 540 (EvoSVM), and 600 (cNB), ameloblastoma data.
We randomly selected partitions for the cross-validations and stratified to ensure that the percentage of eligible studies was the same across partitions. Further, we used the same random seed to ensure that partitions were equivalent when comparing classifiers. For each fold in a 10-fold cross-validation, we trained a classifier on 90% of the training data given a particular combination of parameters in the optimization grid, and assessed performance on the remaining 10%. Because cross-validations are iterative, performance measures were means of 10 values.
To develop a reasonable series of IG thresholds, we inspected a plot of normalized IG weights for BOW extracted from FULL ameloblastoma citations. The absolute values ranged from 0.0 to 1.0, with ameloblastoma having the largest weight; most values were less than 0.20. Thus, our series of threshold values included the following: none (no feature selection), 0.0001, 0.04, 0.08, 0.12, and 0.16. Based on the ameloblastoma results, the thresholds for the influenza data were none and 0.0001.
To ensure feasibility of the EvoSVM optimization runs, we conducted scoping analyses to select appropriate parameter values. By scoping, we mean that we conducted grid optimization with simple validation using a 1:1 split of the training set with no feature selection. However, for mutation types (Gaussian, switching, and sparsity) we followed the methods of phase I in addition to optimization with simple validation. Given the guidance of [43, 44], we considered various values for C and gamma; the default for gamma=1.0 was best. We also confirmed that the default value for epsilon=0.1 was reasonable for our data. We chose a nonlinear kernel based on our pilot study [45]. Given these preliminary analyses, we used the following settings: radial kernel; Gaussian mutation; gamma=1.0; epsilon=0.1; population size=1, 10, 20; and C=1, 10, 20. Thus, population size and C were the input parameter values for the grid optimization in phase II.
For cNB, the input parameter values included smoothing values = 0.001, 0.4, 0.6, 0.8, 1.0 and normalized class weights=false, true, based on [41].
For k-NN, the input parameter values included number of neighbors k = 1, 3, 5, 7 and weighted vote = false, true. Note that when k=1, vote is not relevant.
In addition to cross-validation, we independently validated the best model for each classifier on a reserved test set. Note that the independent tests are stricter than the tests on held-out partitions during cross-validation because data for the former are not used when training classifiers. Thus, the independent tests are probably better estimates of generalizability.
For phase II, we expected that optimization would improve performance for all classifiers. We further expected that after optimization at least one classifier would return recall greater than or equal to 95% with precision greater than 7% for the ameloblastoma data and greater than 6% for the influenza data. Based on the results from phase I, we expected that enriching the feature set extracted from FULL citations with 2G title features would improve performance for cNB.
3. Results
3.1 Phase I (no optimization)
Table 1 displays the independent test results for phase I. In general, there appears to be a complex interaction between classifier, citation portion, and feature set.
Table 1.
Independent test results by classifier, feature type, and citation portion: ameloblastoma dataa
| Recall (%) |
Precision (%) |
F3 (%) |
Recall (%) |
Precision (%) |
F3 (%) |
Recall (%) |
Precision (%) |
F3 (%) |
|
|---|---|---|---|---|---|---|---|---|---|
| BOW / FULL | BOW / TIABS | BOW / TITLES | |||||||
| 1-NN | 69.23 | 57.45 | 67.84 | 64.10 | 60.98 | 63.77 | 46.15 | 45.00 | 46.03 |
| NB | 10.26 | 80.00 | 11.24 | 15.38 | 54.55 | 16.57 | 56.41 | 37.93 | 53.79 |
| cNB | 25.64 | 76.92 | 27.47 | 35.90 | 63.64 | 37.54 | 66.67 | 22.22 | 55.56 |
| EvoSVM | 82.05 | 20.51 | 63.11 | 64.10 | 18.66 | 51.55 | 71.79 | 18.30 | 55.55 |
| 2G / FULL | 2G / TIABS | 2G / TITLES | |||||||
| 1-NN | 61.54 | 60.00 | 61.38 | 64.10 | 58.14 | 63.45 | 35.90 | 35.90 | 35.90 |
| NB | 7.69 | 100.00 | 8.47 | 10.26 | 100.00 | 11.27 | 38.46 | 39.47 | 38.56 |
| cNB | 10.26 | 100.00 | 11.27 | 17.95 | 87.50 | 19.50 | 69.23 | 30.68 | 61.50 |
| EvoSVM | 66.67 | 40.00 | 62.50 | 51.28 | 46.51 | 50.79 | 64.10 | 25.00 | 55.43 |
| 3G / FULL | 3G / TIABS | 3G / TITLES | |||||||
| 1-NN | 61.54 | 63.16 | 61.70 | 64.10 | 62.50 | 63.94 | 28.21 | 28.95 | 28.28 |
| NB | 17.95 | 100.00 | 19.55 | 12.82 | 100.00 | 14.04 | 66.67 | 24.53 | 56.90 |
| cNB | 7.69 | 100.00 | 8.47 | 15.38 | 100.00 | 16.80 | 53.85 | 28.00 | 49.30 |
| EvoSVM | 64.10 | 48.08 | 62.03 | 48.72 | 55.88 | 49.35 | 69.23 | 19.29 | 54.99 |
BOW=bag of words; 2G=2-term n-grams; 3G=3-term n-grams; FULL=title, abstract, metadata; TIABS=title, abstract; 1-NN=1 nearest neighbor; NB=naïve Bayes; cNB=complement naïve Bayes; EvoSVM=evolutionary support vector machine
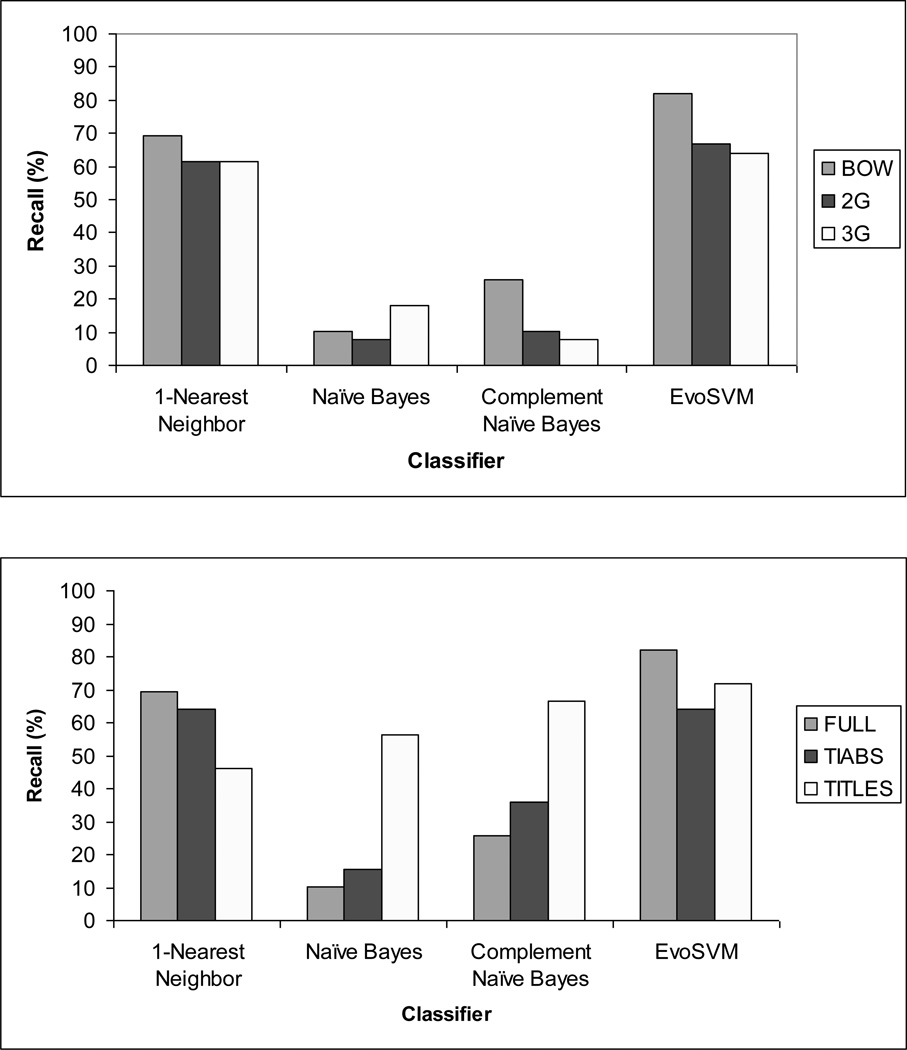
Over 9 possible conditions (3 citation portions × 3 feature sets), EvoSVM returned the best recall (82.05%) for BOW extracted from FULL citations; 1-NN returned the best F3 (67.84%), also for BOW extracted from FULL citations. NB and cNB returned the worst recall (7.69%) and F3 (8.47%) for 2G and 3G extracted from FULL citations.
Over all conditions, recall was best for EvoSVM 5 of 9 times. Precision was maximal when recall was very low, e.g., precision=100% and recall=7.69% for NB, 2G, FULL. NB was the weakest classifier regarding F3 (range: 8.47% to 56.90%).
Fig. 2 (top) displays the results for recall as a function of classifier and feature set when features were extracted from the FULL citation. Using BOW appears to improve recall for EvoSVM, 1-NN, and cNB, but not for NB.
Fig. 2.
Recall varied with classifier and feature set (top), as well as citation portion (bottom). BOW=bag of words; 2G=2-term n-grams; 3G=3-term n-grams; FULL=title, abstract, and metadata; TIABS= title and abstract; ameloblastoma data.
Fig. 2 (bottom) displays the results for recall as a function of classifier and citation portion when the feature set was BOW. Metadata in the FULL citation appear to improve recall for EvoSVM and 1-NN, but not for NB and cNB. (Consider that the difference between FULL and TIABS is the metadata in FULL.) However, extracting BOW from TITLES was associated with best recall for cNB and NB, and was second to FULL citations for EvoSVM.
No classifier reached the recall criterion of at least 95% for acceptable performance.
3.2 Phase II (optimization with cross-validation)
Based on phase I results, we dropped NB from further consideration. We optimized features and parameters with respect to recall and cross-validated models for k-NN, cNB, and EvoSVM using BOW extracted from FULL citations. We also cross-validated optimized models on enriched feature sets (BOW plus 2G title features). All independent tests applied the best training models from the grid optimizations with cross-validations to the reserved data. The best feature-parameter combinations per classifier were the same across ameloblastoma and influenza datasets.
Tables 2, 3, and 4 display the results for optimization with cross-validation; recall and precision are in bold for models that surpassed both cutoffs. Table 5 displays the independent test results.
Table 2.
Classifier performance after grid optimization with 10-fold cross-validation: ameloblastoma dataa
| IG thresholdb | Best parameter set | MN recall (SD) % | MN precision (SD) % | MN F3 (SD) % |
|---|---|---|---|---|
| EvoSVMc | ||||
| none | C=20, pop size=10 | 89.46 (11.05) | 14.01 (2.64) | 58.15 (10.20) |
| 0.0001 | C=1, pop size=10 | 100.00 (0.00) | 13.11 (1.37) | 60.14 (5.58) |
| 0.04 | C=1, pop size=10 | 87.14 (9.71) | 22.31 (5.90) | 67.52 (14.52) |
| 0.08 | C=10, pop size=10 | 80.54 (14.03) | 22.50 (6.16) | 64.02 (14.65) |
| 0.12 | C=10, pop size=10 | 79.11 (15.53) | 29.24 (6.51) | 67.58 (13.77) |
| 0.16 | C=10, pop size=10 | 79.11 (15.53) | 36.51 (7.87) | 70.84 (13.27) |
| cNBd | ||||
| none | smoothing value=.001, normalized=true | 97.32 (5.37) | 8.72 (0.86) | 48.27 (4.53) |
| 0.0001 | smoothing value=.4, normalized=false | 74.82 (16.74) | 39.54 (9.56) | 68.69 (15.36) |
| 0.04 | smoothing value=.4, normalized=false | 88.21 (10.93) | 33.79 (6.23) | 75.97 (11.50) |
| 0.08 | smoothing value=.001, normalized=false | 80.36 (12.29) | 32.50 (7.97) | 70.04 (14.29) |
| 0.12 | smoothing value=.6, normalized=false | 80.36 (15.65) | 24.72 (5.04) | 65.60 (12.38) |
| 0.16 | smoothing value=.001, normalized=true | 71.25 (12.31) | 38.98 (12.18) | 65.80 (15.16) |
| k-NNe | ||||
| none | k=1 | 52.78 (12.35) | 52.02 (11.69) | 52.70 (7.04) |
| 0.0001 | k=1 | 32.86 (11.82) | 47.30 (23.35) | 33.90 (11.87) |
| 0.04 | k=1 | 31.07 (15.45) | 41.05 (16.66) | 31.84 (13.30) |
| 0.08 | k=1 | 38.04 (15.61) | 47.99 (23.03) | 38.84 (14.99) |
| 0.12 | k=1 | 47.32 (13.95) | 58.33 (13.59) | 48.23 (10.44) |
| 0.16 | k=5, weighted vote=true | 44.64 (13.74) | 12.15 (3.39) | 35.22 (9.85) |
Bag of words extracted from full citations.
IG=information gain; feature set size varies inversely with IG threshold.
EvoSVM = evolutionary support vector machine; radial kernel; Gaussian mutation; gamma=1.0; epsilon=0.1; C=1, 10, 20; population size=1, 10, 20.
cNB= complement naïve Bayes; smoothing values=.001, .4, .6, .8, 1.0; normalized class weights=true, false.
k-NN=k-nearest neighbor; k=1, 3, 5, 7 neighbors; weighted vote=true, false, n/a when k=1; cosine similarity measures.
Table 3.
Classifier performance after 10-fold cross-validation: enriched feature set, ameloblastoma dataa
| IG thresholdb | Best parameter set | MN recall (SD) % | MN precision (SD) % | MN F3 (SD) % |
|---|---|---|---|---|
| EvoSVMc | ||||
| 0.0001 | C=1, pop size=10 | 100.00 (0.00) | 14.41 (1.87) | 62.74 (7.01) |
| cNBd | ||||
| none | smoothing value=.001, normalized=true | 100.00 (0.00) | 10.96 (1.21) | 55.18 (5.51) |
| k-NNe | ||||
| none | k=1 | 41.79 (18.04) | 49.69 (20.78) | 42.46 (15.99) |
Bag of words extracted from full citations plus overweighted titles.
IG=information gain; number of features = 1677 when IG >=0.0001; 4607 features when IG threshold=none.
EvoSVM=evolutionary support vector machine; radial kernel; Gaussian mutation; gamma=1.0; epsilon=0.1; C=1, 10, 20; population size=1, 10, 20.
cNB=complement naïve Bayes; smoothing values=.001, .4, .6, .8, 1.0; normalized class weights=true, false.
k-NN=k-nearest neighbor; k=1, 3, 5, 7 neighbors; weighted vote=true, false, n/a when k=1; cosine similarity measures.
Table 4.
Classifier performance after optimization and validation: influenza data
| Grid optimization with 10-fold cross-validationa | |||||
|---|---|---|---|---|---|
| IG thresholdb | N features | Best parameter set | MN recall (SD) % | MN precision (SD) % | MN F3 (SD) % |
| EvoSVMc | |||||
| none | 6828 | C=20, pop size=20 | 74.72 (10.37) | 7.72 (1.00) | 40.00 (5.17) |
| 0.0001 | 2205 | C=1, pop size=10 | 100.00 (0.00) | 10.69 (0.65) | 54.48 (3.02) |
| cNBd | |||||
| none | 6828 | smoothing value=.001, normalized =true | 97.64 (3.12) | 7.14 (0.24) | 43.06 (1.42) |
| 0.0001 | 2205 | smoothing value=.4, normalized=false | 69.56 (8.71) | 41.02 (7.09) | 65.04 (8.80) |
| k-NNe | |||||
| none | 6828 | k=1 | 36.70 (10.51) | 34.82 (9.42) | 36.50 (9.79) |
| 0.0001 | 2205 | k=1 | 24.79 (7.38) | 36.73 (13.59) | 25.62 (7.64) |
| 10-fold cross-validation: enriched feature setf | |||||
| IG threshold | N features | Best parameter set | MN recall (SD) % | MN precision (SD) % | MN F3 (SD) % |
| EvoSVM | |||||
| 0.0001 | 2913 | C=1, pop size=10 | 100.00 (0.00) | 10.74 (0.59) | 54.61 (2.73) |
| cNB | |||||
| none | 9752 | smoothing value=.001, normalized=true |
99.52 (1.43) | 7.48 (0.27) | 44.62 (1.51) |
| k-NN | |||||
| none | 9752 | k=1 | 36.73 (13.92) | 31.86 (9.09) | 36.18 (11.35) |
Bag of words extracted from full citations.
IG=information gain.
EvoSVM=evolutionary support vector machine; radial kernel; Gaussian mutation; gamma=1.0; epsilon=0.1; C=1, 10, 20; population size=1, 10, 20.
cNB=complement naïve Bayes; smoothing values=.001, .4, .6, .8, 1.0; normalized class weights=true, false.
k-NN=k-nearest neighbor; k=1, 3, 5, 7 neighbors; weighted vote=true, false, n/a when k=1; cosine similarity measures.
Bag of words extracted from full citations plus overweighted titles.
Table 5.
Independent test results by classifier for influenza and ameloblastoma dataa
| Evolutionary support vector machine |
Complement naïve Bayes | k-nearest neighbor | |
|---|---|---|---|
| Recall (%) | |||
| Full citationb | |||
| Influenza | 79.44 | 97.20 | 29.91 |
| Ameloblastoma | 100.00 | 97.44 | 69.23 |
| Full citation + weighted titlesc | |||
| Influenza | 90.65 | 98.13 | 25.23 |
| Ameloblastoma | 100.00 | 94.87 | 58.97 |
| Mean rankd, e | 2.5 | 2.5 | 1.0 |
| Precision (%) | |||
| Full citation | |||
| Influenza | 8.20 | 7.30 | 30.48 |
| Ameloblastoma | 13.09 | 9.22 | 57.45 |
| Full citation + weighted titles | |||
| Influenza | 8.90 | 7.58 | 25.47 |
| Ameloblastoma | 13.40 | 10.95 | 60.53 |
| Mean ranke | 2.0 | 1.0 | 3.0 |
| F3 (%) | |||
| Full citation | |||
| Influenza | 42.51 | 43.56 | 29.97 |
| Ameloblastoma | 60.10 | 49.80 | 67.84 |
| Full citation + weighted titles | |||
| Influenza | 47.25 | 44.71 | 25.25 |
| Ameloblastoma | 60.74 | 53.71 | 59.12 |
| Mean rankf | 2.5 | 1.8 | 1.8 |
Bag of words extracted from full citations.
Bag of words extracted from full citations plus overweighted titles.
Higher ranks associated with better performance.
Mean ranks significantly different for recall and precision: Friedman chi2 (2 df)=6, P=.0498 and Friedman chi2 (2 df)=8, P=.0183, respectively.
Mean ranks not significantly different for F3: Friedman chi2 (2 df)=1.5, P= .4724.
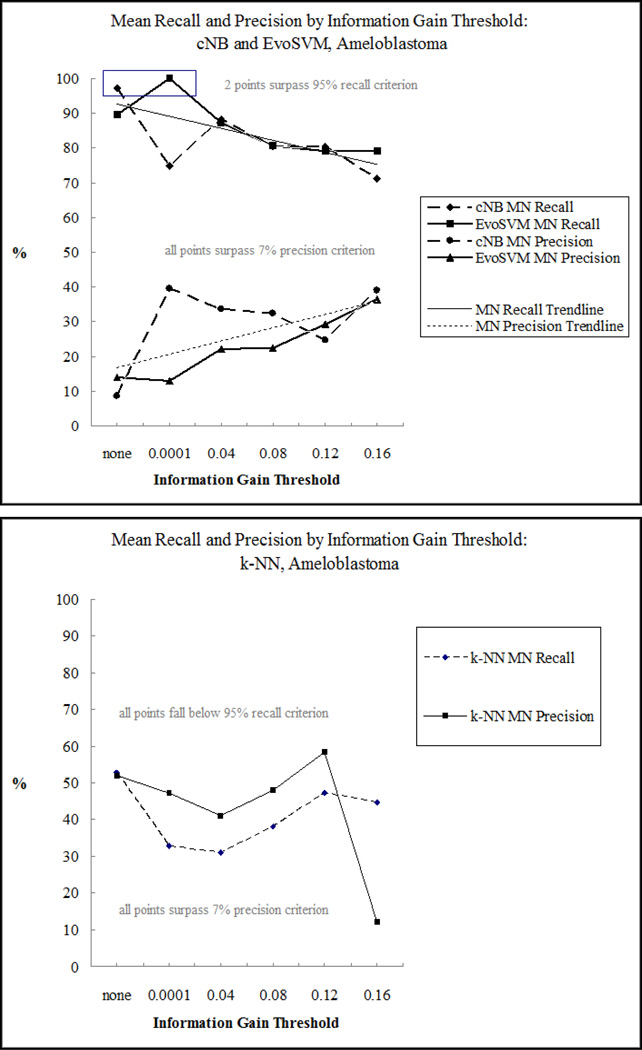
Fig. 3 displays mean recall and precision as a function of IG threshold for the ameloblastoma data. The curves for both cNB and EvoSVM were inversely related, which is typical of the tradeoff between recall and precision. Two points surpassed both recall and precision criteria: when the IG threshold=none for cNB and 0.0001 for EvoSVM. For k-NN, the curves were similar, but diverged for the largest IG threshold. Although k-NN always surpassed the precision cutoff, it never met the recall criterion.
Fig. 3.
Mean recall and precision varied by information gain (IG) threshold. Complement naïve Bayes (cNB) and evolutionary support vector machine (EvoSVM) surpassed both recall and precision cutoffs when all features were selected or when the IG weight was >= 0.0001 (top). For k-nearest neighbor (k-NN), no points surpassed the recall cutoff (bottom).
3.2.1 EvoSVM
The best model for EvoSVM over all IG thresholds involved a subset of features for which the IG weight was >= 0.0001; n=1430 (40%) and n=2205 (32%) features, ameloblastoma and influenza data, respectively. The best parameter set was C=1 and population size=10 (see Tables 2 and 4).
For the independent tests with ameloblastoma data, recall was stable when compared to the best optimization results, i.e., recall=100% for both BOW and enriched BOW (see Tables 2, 3, and 5). However, with influenza data, recall degraded from 100% to 79.44% and 90.65% for BOW and the enriched BOW, respectively (see Tables 4 and 5). Enrichment boosted precision 2.4% (13.40% vs. 13.09%, ameloblastoma) and 8.5% (8.90% vs. 8.20%, influenza) (see Table 5). We computed the percentage improvement as [(.0890–.0820)/.0820] × 100=8.5%. EvoSVM surpassed both recall and precision thresholds for the ameloblastoma data, and the precision threshold for influenza data. However, it failed with respect to recall for the influenza data.
Compared to the results from phase I, recall for the optimized model on the ameloblastoma test set was 21.9% better (100% vs. 82.05%) and F3 was 3.8% worse (60.74% vs. 63.11%). (See Table 1, BOW/FULL and Table 5.)
3.2.2 cNB
The best optimized model for cNB over all IG thresholds involved the full set of features: n=3574 and n=6828, ameloblastoma and influenza data, respectively. The best parameter set was smoothing value=0.001 and normalized weights for each class (see Tables 2 and 4).
For the independent tests, recall was relatively stable when compared to the best optimization results (ameloblastoma and influenza data) (see Tables 2–5). Enrichment boosted precision 18.8% (10.95% vs. 9.22%, ameloblastoma) and 3.8% (7.58% vs. 7.30%, influenza) (see Table 5). cNB surpassed both recall and precision thresholds with influenza data (both feature sets) and ameloblastoma data (BOW). However, it just missed the recall threshold of 95% with ameloblastoma data and the enriched BOW (recall=94.87%).
Compared to the results from phase I, recall for the optimized model on the ameloblastoma test set was 2.8 times better (97.44% vs. 25.64%); F3 was 81.3% better (49.80% vs. 27.47%). (See Table 1, BOW, FULL and Table 5).
3.2.3 k-NN
The best optimized model for k-NN over all IG thresholds was based on the full set of features. The best parameter setting was k=1, vote not applicable (see Tables 2 and 4).
The results for the independent tests were quite mixed. For example, recall improved when compared to the best optimization results for the ameloblastoma data, but degraded for the influenza data (see Tables 2, 4, and 5). Enrichment boosted precision for the ameloblastoma data, but degraded precision for the influenza data (see Table 5). k-NN failed to meet the recall threshold for both datasets regardless of feature set, whereas it always surpassed the precision threshold.
For the ameloblastoma data, the results of the independent test for BOW extracted from FULL citations were the same as in phase I (see Table 1, BOW/FULL and Table 5). This is because the models were the same.
3.2.4 Comparison of classifiers
Following the advice of Demsar [46], we computed an omnibus Friedman test statistic to assess differences among mean ranks for 3 classifiers per performance measure (see Table 5). The Friedman test is a robust, nonparametric alternative to repeated measures ANOVA. When the Friedman test statistic was statistically significant (P < .05), we computed Bonferroni-Dunn tests for post hoc comparisons; we adjusted alpha for the number of comparisons to control the Type I error rate. Note that higher ranks are associated with better performance.
Mean ranks for recall were significantly different: 2.5 (EvoSVM), 2.5 (cNB), and 1.0 (k-NN); Friedman chi2 (2 df) = 6, P =.0498. Because the post hoc comparison for EvoSVM vs. k-NN was the same as for cNB vs. k-NN—2.5 vs. 1.0—alpha was not adjusted. For EvoSVM or cNB vs. k-NN, mean recall was significantly different: z = 2.12, P = .034. Thus, recall was not significantly different for EvoSVM vs. cNB, but was when each was compared to k-NN. Recall was always better for EvoSVM or cNB vs. k-NN. Overall, recall usually improved or was stable when features were enriched by overweighting titles for EvoSVM and cNB, but not for k-NN.
The mean ranks for precision were significantly different: 2.0 (EvoSVM), 1.0 (cNB), and 3.0 (k-NN); Friedman chi2 (2 df) = 8, P = .0183. Because 3 post hoc comparisons were computed, the adjusted alpha = .05/3 = .0167. For EvoSVM vs. cNB: z = 1.41, P = .1585. For EvoSVM vs. k-NN, z = −1.41, P = .1585. For cNB vs. k-NN: z = −2.83, P=.0047. Thus, precision was not significantly different for EvoSVM vs. cNB or EvoSVM vs. k-NN, but was for cNB vs k-NN. Precision was always better for k-NN when compared to cNB. In general, precision improved for EvoSVM and cNB when features were enriched by overweighting titles, whereas results for k-NN were mixed.
The mean ranks for F3 were not significantly different: 2.5 (EvoSVM), 1.8 (cNB), and 1.8 (k-NN); Friedman chi2 (2 df) = 1.5, P = .4724. Because the omnibus test was not statistically significant, post hoc comparisons were not warranted.
4. Discussion
4.1 Implications
To understand the implications of this research, consider the following scenario. Assume that (1) a reliable machine learning system exists to assist systematic reviewers when screening citations; (2) 3000 citations have been retrieved; (3) human reviewer(s) complete the first pass through the entire set of citations and label 180 (6%) as eligible for full-text review; and (4) two machine learning classifiers are available (EvoSVM and cNB). Given sampling variability, our best estimates for recall and precision are the averages for the independent test results on the enriched feature sets. Thus, further assume that recall and precision are 95.32% and 11.15% for EvoSVM, and 96.50% and 9.27% for cNB (based on Table 5).
The questions of concern to potential users are: How many citations will each machine learning classifier identify as eligible and how does this compare to screening the entire set once again? If the system is useful, reviewers need not consider further the disproportionately large number of citations labeled as ineligible both by human(s) and machine.
If the reviewers choose EvoSVM, the classifier correctly labels 172 citations and incorrectly labels another 1443 as eligible. Thus, a noisy set of 1615 true and false positives (172+1443=1615) is returned for the second pass through the citations by at least one more human reviewer—we refer to the size of this set as the number needed to screen (NNS). However, the NNS should be adjusted somewhat by the 8 citations overlooked by the machine, but identified by human(s). This is because recall is not perfect. Thus, the NNS for EvoSVM is 1615+8=1623, which is a 46% reduction in the size of the initial retrieval set: (3000-1623)/3000=.459.
If the reviewers choose cNB, the classifier correctly labels 174 citations and incorrectly labels another 1768 as eligible. A set of 1942 true and false positives is returned. Adding in the 6 citations overlooked by the machine, the NNS is 1948, which is a 35% reduction in the size of the initial retrieval set.
Clearly, if a reliable system were in place and both classifiers were reasonably efficient, systematic reviewers would choose EvoSVM in favor of cNB because the NNS=1623 for EvoSVM and 1948 for cNB. Nevertheless, until we have more citations from SRs on topics where NR studies are likely, our estimates for recall and precision may be unrealistic.
A major challenge for future research is boosting precision to reduce further the screening burden while maintaining very high recall. More than likely, we need feature sets that capitalize on both the structure of citations and the language that scientists and indexers use to describe studies. Regarding the latter, review teams outside of the United States are likely to search EMBASE, which is the European counterpart of MEDLINE. However, indexers use different terms for the same concepts, and MeSH and EMTREE terms can appear in different places in the citation. Thus, modeling structure is a challenge if we want to extract indexing terms and tag for source. In this paper, we demonstrated that adding contextual information from pairs of title words tends to boost precision modestly—suggesting that we can do a better job of modeling the format and scientific language of biomedical citations.
4.2 Classifier performance
The results were somewhat surprising. For phase I, we had expected that without optimization, recall and overall performance would be best using 2- or 3-term n-grams extracted from complete citations. Instead, using single processed words (BOW) from FULL citations was associated with best performance. This suggests that indexing in the complete citation improves performance, even when the indexing terms are processed as single words. To improve this feature set in future work, we could preserve the MeSH and EMTREE terms (phrases), which would yield a feature set similar to the one used by Cohen and colleagues [23, 47].
Because none of the classifiers from phase I attained high enough recall to be of use, optimization in phase II was warranted.
For phase II, we had expected that all classifiers would benefit from optimization. This was generally true for EvoSVM and cNB, but not for k-NN. As it turned out, the optimized model for k-NN was the same as the one we used during phase I. Additionally, just EvoSVM benefited from selecting features based on IG. The results did support our expectation that, with optimization, one or more classifiers would return recall at least as high as 95% and precision greater than 6% or 7%, depending on the dataset. Both EvoSVM and cNB met these criteria, but generalization performance for EvoSVM was not as good as for cNB. This suggests either sampling variability or overfitting of EvoSVM during training. If the latter, the parameter C may not have been tuned well because C purportedly controls overfitting ([35], p. 301). Additionally, a radial kernel may have been inappropriate (see below).
Additionally, we had expected that enriching the BOW from full citations by overweighting titles would improve performance for cNB. It was somewhat surprising that enrichment improved performance for both cNB and EvoSVM.
Although researchers currently favor variants of both of these classifiers [22–25], the evidence suggests that optimization is necessary to boost performance. In fact, the results for cNB were startling with an almost three-fold improvement for recall and an 81% improvement for overall performance when comparing phase I and phase II results for ameloblastoma data.
4.3 Limitations
The major limitation of this study is that the citations came from just two systematic reviews. Future comparative studies of classifiers should use citations from several reviews, paying attention to phrases for NR study designs that meet inclusion criteria as specified in the protocols. Presumably, more precise classification is possible for randomized controlled trials because the indexing is better than for NR studies (see the Introduction here and in [45]).
Another limitation is that we wrapped feature selection around grid optimization of classifier parameters, ignoring the class imbalance problem [48]. While using a wrapper strategy is a well-known approach [49], a better one could involve selecting features within the positive (include) and negative (exclude) classes before grid optimization (e.g., see [49, 50]). Recently, Le and colleagues [51] compared other optimization methods, including stochastic gradient descent (SGD), limited memory BFGS (L-BFGS), and conjugate gradient (CG) methods. They reported that in contrast to the favored SGD method, L-BFGS and CG methods outperform SGD with respect to speed and accuracy. However, their overall conclusion was that performance of the optimization method varies with the research problem.
Certainly, a more thorough comparison of parameter settings for EvoSVM is required as this classifier has quite a few parameters. In particular, a study comparing performance as a function of kernel is essential in the context of classifying biomedical citations. Because generalization performance is “dominated by the chosen kernel function” ([52], p. 1313), researchers are developing automatic methods for learning kernel functions. A promising nonparametric approach was described in [52], wherein a family of simple nonparametric kernel learning (NPKL) algorithms was presented. Simple NPKL algorithms are reportedly as accurate as other NPKL methods, but more efficient and scalable. This line of research is timely inasmuch as parametric SVMs do not scale well for many applications, selection of the appropriate kernel is not obvious, and parametric kernels may be inappropriate for this task.
Although the results from our study and [45] suggest that EvoSVM with a nonlinear kernel is promising, the runtimes are much longer than for cNB. In the near term, cNB may be the better choice to semi-automate citation screening, especially when the number of citations is large. Finally, in our opinion, conditional random fields [53] and latent Dirichlet allocation [54] might profitably be compared to variants of cNB and SVM.
5. Conclusion
We have demonstrated that machine learning classifiers can help identify NR studies eligible for full-text screening by systematic reviewers. We have further shown that optimization can markedly improve classifier performance. In our opinion, careful comparative research is needed before a classifier is chosen to semi-automate screening citations. Further, stability of performance for optimized classifiers needs to be demonstrated over various medical review topics.
Acknowledgements
We thank the anonymous reviewers for their thoughtful remarks. Additionally, we thank Drs. Thankam Thyvalikakath and Richard Oliver for help with labeling ameloblastoma citations; Ms. Anne Littlewood, Cochrane Oral Health Group Trials Search Coordinator, and Ms. Jill Foust, Reference Librarian, University of Pittsburgh Health Sciences Library System, for help with database searches; and Mr. Richard Wilson and Mr. Eugene Tseytlin for technical assistance. This research was supported, in part, by the Pittsburgh Biomedical Informatics Training Program (5 T15 LM/DE07059) and an NIH award from the US National Library of Medicine (K99LM010943).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bastian H, Glasziou P, Chalmers I. Seventy-five trials and eleven systematic reviews a day: how will we ever keep up? PLoS Med. 2010;7 doi: 10.1371/journal.pmed.1000326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cohen AM, Adams CE, Davis JM, Yu C, Yu PS, Meng W, et al. Evidence-based Medicine, the Essential Role of Systematic Reviews, and the Need for Automated Text Mining Tools. IHI'10. Arlington, Virginia, USA: ACM; 2010. pp. 376–380. 978-1-4503-0030-8/10/11. [Google Scholar]
- 3.Cochrane Collaboration Methods Groups. [Accessed: 7 August 2011]; http://www.cochrane.org/contact/methods-groups. [Google Scholar]
- 4.Cochrane Collaboration. [Accessed: 7 August 2011]; http://www.cochrane.org/ [Google Scholar]
- 5.Agency for Healtcare Research and Quality (AHRQ) Evidence-based Practice Centers. [Accessed: 7 August 2011]; http://www.ahrq.gov/clinic/epc/
- 6.Atkins D, Fink K, Slutsky J. Better Information for Better Health Care: The Evidence-based Practice Center Program and the Agency for Healthcare Research and Quality. Annals of Internal Medicine. 2005;142:1035–1041. doi: 10.7326/0003-4819-142-12_part_2-200506211-00002. [DOI] [PubMed] [Google Scholar]
- 7.Tricco AC, Brehaut J, Chen MH, Moher D. Following 411 Cochrane protocols to completion: a retrospective cohort study. PLoS One. 2008;3:e3684. doi: 10.1371/journal.pone.0003684. PMID18997866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aphinyanaphongs Y, Tsamardinos I, Statnikov A, Hardin D, Aliferis CF. Text categorization models for high-quality article retrieval in internal medicine. J Am Med Inform Assoc. 2005;12:207–216. doi: 10.1197/jamia.M1641. PMID15561789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kilicoglu H, Demner-Fushman D, Rindflesch TC, Wilczynski NL, Haynes RB. Towards automatic recognition of scientifically rigorous clinical research evidence. J Am Med Inform Assoc. 2009;16:25–31. doi: 10.1197/jamia.M2996. PMID18952929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wilczynski NL, Morgan D, Haynes RB. An overview of the design and methods for retrieving high-quality studies for clinical care. BMC Med Inform Decis Mak. 2005;5:20. doi: 10.1186/1472-6947-5-20. PMID15969765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Norris SL, Atkins D. Challenges in Using Nonrandomized Studies in Systematic Reviews of Treatment Interventions. Annals of Internal Medicine. 2005;142:1112–1119. doi: 10.7326/0003-4819-142-12_part_2-200506211-00011. [DOI] [PubMed] [Google Scholar]
- 12.Reeves B, Deeks J, Higgins J, Wells G. Chapter 13: Including Non-randomized Studies. In: Higgins J, Green S, editors. Cochrane Handbook for Systematic Reviews of Interventions. Chichester (UK): Wiley; 2008. [Google Scholar]
- 13.Cohen AM, Hersh WR, Peterson K, Yen PY. Reducing workload in systematic review preparation using automated citation classification. J Am Med Inform Assoc. 2006;13:206–219. doi: 10.1197/jamia.M1929. PMID16357352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McKibbon KA, Wilczynski NL, Haynes RB Hedges Team. Retrieving randomized controlled trials from MEDLINE: a comparison of 38 published search filters. Health Info Libr J. 2009;26:187–202. doi: 10.1111/j.1471-1842.2008.00827.x. PMID19712211. [DOI] [PubMed] [Google Scholar]
- 15.Centre for Reviews and Dissemination. Systematic reviews: CRD's guidance for undertaking reviews in health care. University of York, UK; 2009. [Accessed: 7 August 2011]. http://www.york.ac.uk/inst/crd/index_guidance.htm. [Google Scholar]
- 16.Flemming K, Briggs M. Electronic searching to locate qualitative research: evaluation of three strategies. Journal of Advanced Nursing. 2007;57:95–100. doi: 10.1111/j.1365-2648.2006.04083.x. PMID17184378. [DOI] [PubMed] [Google Scholar]
- 17.Fraser C, Murray A, Burr J. Identifying observational studies of surgical interventions in MEDLINE and EMBASE. BMC Medical Research Methodology. 2006;6:41. doi: 10.1186/1471-2288-6-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leeflang MM, Scholten RJ, Rutjes AW, Reitsma JB, Bossuyt PM. Use of methodological search filters to identify diagnostic accuracy studies can lead to the omission of relevant studies. [Review] Journal of Clinical Epidemiology. 2006:234–240. doi: 10.1016/j.jclinepi.2005.07.014. PMID16488353. [DOI] [PubMed] [Google Scholar]
- 19.Leeflang M, McDonald S, Scholten Rob JPM, Rutjes A, Reitsma Johannes JB. Cochrane Database of Systematic Reviews. Chichester, UK: John Wiley &Sons, Ltd; 2007. Search strategies to identify diagnostic accuracy studies in MEDLINE and EMBASE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shaw R, Booth A, Sutton A, Miller T, Smith J, Young B, et al. Finding qualitative research: an evaluation of search strategies. BMC Medical Research Methodology. 2004;4:5. doi: 10.1186/1471-2288-4-5. PMID15070427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bekhuis T, Thyvalikakath TP, Oliver R. Cochrane Database of Systematic Reviews. issue 4. Chichester, UK: Wiley; 2009. Interventions for treating ameloblastomas of the jaws [protocol] [Google Scholar]
- 22.Frunza O, Inkpen D, Matwin S, Klement W, O'Blenis P. Exploiting the systematic review protocol for classification of medical abstracts. Artificial Intelligence in Medicine. doi: 10.1016/j.artmed.2010.10.005. In Press. [DOI] [PubMed] [Google Scholar]
- 23.Cohen AM, Ambert K, McDonagh M. Cross-topic learning for work prioritization in systematic review creation and update. J Am Med Inform Assoc. 2009;16:690–704. doi: 10.1197/jamia.M3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wallace BC, Trikalinos TA, Lau J, Brodley C, Schmid CH. Semi-automated screening of biomedical citations for systematic reviews. BMC Bioinformatics. 2010;11 doi: 10.1186/1471-2105-11-55. [electronic]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matwin S, Kouznetsov A, Inkpen D, Frunza O. A new algorithm for reducing the workload of experts in performing systematic reviews. Journal of the American Medical Informatics Association. 2010;17:446–453. doi: 10.1136/jamia.2010.004325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Colas F, Brazdil P. Comparison of svm and some older classification algorithms in text classification tasks. International Federation for Information Processing-Publications-IFIP. 2006;217:169. [Google Scholar]
- 27.Cochrane Collaboration. Selecting Studies for your Review: Practicalities of Sifting. [Accessed: 7 August 2011]; http://www.cochrane-net.org/openlearning/HTML/mod8-3.htm. [Google Scholar]
- 28.Jefferson T, Di Pietrantonj C, Al-Ansary LA, Ferroni E, Thorning S, Thomas RE. Cochrane Database of Systematic Reviews. Issue 2. Chichester, UK: Wiley; 2010. Vaccines for preventing influenza in the elderly [updated review] [DOI] [PubMed] [Google Scholar]
- 29.PubMed.gov: US National Library of Medicine, National Institutes of Health. [Accessed: 3 August 2011]; http://www.ncbi.nlm.nih.gov/pubmed/
- 30.EMBASE: Biomedical Answers. [Accessed: 7 August 2011]; http://www.embase.com/ [Google Scholar]
- 31.Rivetti D, Jefferson T, Thomas Roger E, Rudin M, Rivetti A, Di Pietrantonj C, et al. Cochrane Database of Systematic Reviews. Chichester, UK: John Wiley &Sons, Ltd; 2006. Vaccines for preventing influenza in the elderly. [DOI] [PubMed] [Google Scholar]
- 32.Mierswa I, Wurst M, Klinkenberg R, Scholz M, Euler T. YALE (now RapidMiner): rapid prototyping for complex data mining tasks. Proc ACM SIGKDD Int Conf on Knowl Discov Data Mining. 2006 [Google Scholar]
- 33.RapidMiner. [Accessed: 7 August 2011]; http://rapid-i.com/ [Google Scholar]
- 34.RapidMiner Text Plugin. [Accessed: 7 August 2011]; http://sourceforge.net/projects/rapidminer/files/2.%20Text%20Plugin/ [Google Scholar]
- 35.Manning CD, Raghavan P, Schutze H. Introduction to Information Retrieval. New York: Cambridge University Press; 2008. [Google Scholar]
- 36.US National Library of Medicine. National Institutes of Medicine: Medical Subject Headings [MeSH] [Accessed: 3 August 2011]; http://www.nlm.nih.gov/mesh/
- 37.EMTREE: The Life Science Thesaurus. [Accessed: 3 August 2011]; http://embase.com/info/what-is-embase/emtree. [Google Scholar]
- 38.Sebastiani F. Machine learning in automated text categorization. ACM Computing Surveys. 2002;34:1–47. [Google Scholar]
- 39.Lefebvre C, Manheimer E, Glanville J. Cochrane Handbook for Systematic Reviews of Interventions. Chichester, UK: Wiley; 2008. Chapter 6: Searching for Studies. [Google Scholar]
- 40.Hand D, Mannila H, Smyth P. Principles of Data Mining. Cambridge, MA: MIT Press; 2001. Chapter 10: The Naive Bayes Model; pp. 353–356. [Google Scholar]
- 41.Rennie J, Shih L, Teevan J, Karger D. Tackling the poor assumptions of naive bayes text classifiers; Proceedings of the Twentieth International Conference on Machine Learning (ICML); Washington, DC. 2003. [Google Scholar]
- 42.Mierswa I. Evolutionary Learning with Kernels: A Generic Solution for Large Margin Problems; Proc of the Genetic and Evolutionary Computation Conference (GECCO); Seattle, Washington. 2006. [Google Scholar]
- 43.Chow R, Zhong W, Blackmon M, Stolz R, Dowell M. GECCO’08. Atlanta, Georgia: USA ACM; 2008. An efficient SVM-GA feature selection model for large healthcare databases. [Google Scholar]
- 44.Hsu C-W, Chang C-C, Lin C-J. A practical guide to support vector classification. [Accessed: 7 August 2011];2003 :1–12. updated April 2010. http://www.csie.ntu.edu.tw/~cjlin/papers/guide/guide.pdf. [Google Scholar]
- 45.Bekhuis T, Demner-Fushman D. Towards automating the initial screening phase of a systematic review. Stud Health Technol Inform. 2010;160:146–150. PMID 20841667. [PubMed] [Google Scholar]
- 46.Demšar J. Statistical comparisons of classifiers over multiple data sets. The Journal of Machine Learning Research. 2006;7:1–30. [Google Scholar]
- 47.Cohen AM. Optimizing feature representation for automated systematic review work prioritization. AMIA Annu Symp Proc. 2008 PMID18998798. [PMC free article] [PubMed] [Google Scholar]
- 48.Weiss GM. Mining with rarity: a unifying framework. ACM SIGKDD Explorations Newsletter. 2004;6:7–19. [Google Scholar]
- 49.Parsons L, Haque E, Liu H. Subspace clustering for high dimensional data: a review. ACM SIGKDD Explorations Newsletter. 2004;6:90–105. [Google Scholar]
- 50.Zheng Z, Wu X, Srihari R. Feature selection for text categorization on imbalanced data. ACM SIGKDD Explorations Newsletter. 2004;6:80–89. [Google Scholar]
- 51.Le QV, Ngiam J, Coates A, Lahiri A, Prochnow B, Ng AY. On Optimization Methods for Deep Learning. In: Getoor Lise, Scheffer Tobias., editors. Proceedings of the 28th International Conference on Machine Learning (ICML-11); ACM; June 2011; Bellevue, WA, USA. pp. 265–272. [Google Scholar]
- 52.Zhuang J, Tsang IW, Hoi SCH. A Family of Simple Non-Parametric Kernel Learning Algorithms. Journal of Machine Learning Research. 2011;12:1313–1347. [Google Scholar]
- 53.Sutton C, McCallum A. An introduction to conditional random fields for relational learning. In: Getoor Lise, Taskar Ben., editors. Introduction to statistical relational learning. MIT Press; 2006. pp. 95–130. [Google Scholar]
- 54.Blei DM, Ng AY, Jordan MI. Latent dirichlet allocation. The Journal of Machine Learning Research. 2003;3:993–1022. [Google Scholar]