Abstract
Animal toxins are of interest to a wide range of scientists, due to their numerous applications in pharmacology, neurology, hematology, medicine, and drug research. This, and to a lesser extent the development of new performing tools in transcriptomics and proteomics, has led to an increase in toxin discovery. In this context, providing publicly available data on animal toxins has become essential. The UniProtKB/Swiss-Prot Tox-Prot program (http://www.uniprot.org/program/Toxins) plays a crucial role by providing such an access to venom protein sequences and functions from all venomous species. This program has up to now curated more than 5’000 venom proteins to the high-quality standards of UniProtKB/Swiss-Prot (release 2012_02). Proteins targeted by these toxins are also available in the knowledgebase. This paper describes in details the type of information provided by UniProtKB/Swiss-Prot for toxins, as well as the structured format of the knowledgebase.
Keywords: UniProtKB/Swiss-Prot Tox-Prot program, Database, Curation, Venom protein, Animal toxin, Bioinformatics
1. Introduction
Venomous animals have developed a system to capture preys which is quite original: they inject a mixture of toxic components to paralyze or weaken their prey. These toxic components able to impair healthy organisms have been used for centuries by traditional Chinese and Indian medicine to treat some pathological states. More recently, they have been the focus of pharmaceutical research to discover new drug leads. Their potential use in the occidental pharmacopoeia along with the development of new tools in transcriptomics and proteomics, has led to an exponential increase in the rate of venom component discovery (King et al., 2008).
The generation of large quantities of data makes it necessary to develop efficient management systems such as databases to permit a rapid access to specific information. Four databases provide such resource for animal toxins (Jungo et al., 2010). ConoServer (Kaas et al., 2012) and ArachnoServer (Herzig et al., 2011), two specialized databases, offer the community a comprehensive set of curated data on cone snail and spider toxins. The Animal Toxin DataBase (ATDB) (He et al., 2010) restricts its scope to toxins and their targets and provides standardized information gathered from different databases. UniProtKB is a general knowledgebase that provides both manually curated and automatically curated venom proteins from all phyla and their target protein(s) in the two sections UniProtKB/Swiss-Prot and UniProtKB/TrEMBL (Jungo and Bairoch, 2005; Magrane et al., 2011; The UniProt consortium, 2012). These last two databases present an overview of all venom proteins, allowing for comparison of toxins across the different phyla of venomous animals. Both also offer information on the toxins target proteins (ion channels, receptors, hemostasis proteins, etc).
In this paper, we describe the animal toxin annotation in UniProtKB/Swiss-Prot with a special focus on the format and give some insight on the website, both type of information being necessary for users to easily retrieve toxins of interest from UniProtKB.
2. Venom proteins in UniProtKB
The prerequisite for an entry to appear in UniProtKB is the existence of a sequence of consecutive amino-acids even if only fragmentary. Three sources are commonly used: (i) the translations of coding sequences (CDS) submitted to the International Nucleotide Sequence Database Collaboration (INSDC) (also referred as EMBLbank(ENA)/GenBank/DDBJ) (Karsch-Mizrachi et al., 2012), (ii) sequences extracted from the literature and (iii) direct submission of peptide/protein sequences by authors (Fig. 1). New protein sequences obtained by translation of nucleotide sequences together with species and other information are integrated in UniProtKB/TrEMBL and automatically curated. Those accompanied with experimental data are further manually curated and integrated in UniProtKB/Swiss-Prot. Peptide sequences obtained by Edman degradation or MS/MS de novo sequencing are directly integrated and curated into UniProtKB/Swiss-Prot. These sequences are either directly submitted to UniProtKB/Swiss-Prot by authors or typed in from the literature by curators. A major effort is done to curate these sequences that are only published in literature and not submitted to any database.
Figure 1.
Schematic overview of venom protein sequence retrieval and annotation process.
2.1 Sequence
It is UniProtKB/Swiss-Prot policy to describe the product(s) of one gene per entry. While this policy is easy to follow for organisms which have their genome sequenced, its application to venom proteins is not trivial and gene assignment is hampered by the lack of knowledge on genomes and by the number of gene duplications (Kordis et al., 2000). As a consequence, closely related sequences are generally kept separate when precise reliable gene assignment is not possible.
A corollary of the ‘one gene-one entry’ rule is that different proteins/peptides encoded by the same gene are all described in the same entry (e.g. UniProt ID: P30403). The same holds true for alternative splicing products (e.g. UniProt ID: P0CI42) and mRNA editing products. The information about mature peptide/protein length and positions is contained in the ‘Sequence annotation’ field, also named ‘Features’. Another rule concerning sequences in UniProtKB is to show the longest protein sequence known. ‘Molecular processing’ information (such as signal sequence, propeptide and mature peptide) is hence indicated on the basis of the longest sequence shown.
Sequence fragments are accepted in UniProtKB (e.g. UniProt ID: P0C8I8), as well as sequences containing uncertain residues. Leucine and isoleucine residues, indistinguishable in MS/MS de novo sequenced proteins are tagged with the topic ‘Unsure’ in the ‘Sequence annotation’ field (e.g. UniProt ID: P86261). There are however only few such cases since proteomic analyses are often done in parallel with transcriptomic/genomic studies (Escoubas et al., 2006; Gowd et al., 2008; Jakubowski et al., 2004).
2.2 General information
Information is dispersed in the scientific literature and in some specialized databases. As a universal protein knowledgebase, UniProtKB/Swiss-Prot combines detailed information from multiple sources, and presents this in a structured manner in the ‘General annotation (Comments)’ field. The most interesting information about animal toxin, in addition to the sequence and PTMs, consists of its function and its biological activity (both stored in the ‘Function’ topic, see next paragraph). However, supplementary information is also of interest. This includes but is not limited to: measures of the biological activity (lethal, paralytic and effect doses) (stored in the ‘Toxic dose’ topic), the monomeric or multimeric state of the protein (‘Subunit structure’ topic), the domain(s) present in the protein (‘Domain’ topic), the mass experimentally measured (‘Mass spectrometry’ topic), the similarity of sequence (family) with other proteins (‘Sequence similarities’ topic), and the tissue that produces the toxin (‘Tissue specificity’ topic). The tissue is always either venom gland or venom duct, except for sea anemones. This information is of importance, since it is one of the keys to retrieve the venom protein(s) of interest. Two other topics should be mentioned, since they provide information impossible to indicate elsewhere (‘Miscellaneous’ and ‘Caution’ topics) (e.g. UniProt ID: P0CC13 and Q9GQW3).
2.2.1 Toxin function, and biological activity
Studies of venom action have led to the development of numerous therapeutic applications as well as improving our understanding of the basis of physiological processes. The well-known example of snake venom that drastically lowers the blood pressure in human victims has led, for example, to the development of captopril to treat hypertension as well as some types of congestive heart failure (Koh and Kini, 2012). As a second example, the venom from Texas coral snake, the bite of which produces intense and unremitting pain, is studied to probe molecular mechanisms underlying pain sensation (Bohlen et al., 2011). The development of new technologies has permitted to very precisely associate venom effects with specific protein(s) (e.g UniProt ID: G9I929 and G9I930 for the toxin responsible of pain sensation cited above). More data is thus available on new toxins and their function.
To facilitate access to such knowledge, UniProtKB/Swiss-Prot describes venom protein activity in both human- and computer-readable forms. Precise molecular targets and biological activities are indicated in the ‘General annotation’ field under the ‘Function’ topic as free-text (e.g. UniProt ID: P26349). The actual ion channels/receptors of the prey that are targeted by a given toxin are described using two official nomenclatures, i.e. the International Union of Basic and Clinical Pharmacology nomenclature (IUPHAR) (Alexander et al., 2011) and HUGO Gene Nomenclature Committee (HGNC) (Seal et al., 2011). The IUPHAR nomenclature is provided to respond to the needs of neuroscientists, pharmacologists and toxinologists who commonly use this nomenclature, whereas the HGNC nomenclature provides official short gene names that allow a consistent and rapid retrieval of all targets available in separate UniProtKB entries. In addition to ion channels/receptors, this nomenclature allows to retrieve other targets that are not classified by the IUPHAR nomenclature, such as blood coagulation factors largely modulated by snake toxins, for example.
General function and biological activities are also described in a standardized computer-parsable format through the use of keywords. For example, all toxins that act by interfering with calcium currents can be retrieved by querying ‘Keyword: “Calcium channel inhibitor”’ in the search tool of the UniProt website. To have more precise and hierarchically related terms, as well as to facilitate data integration with other resources, we plan to use the Gene Ontology (GO) terms to describe the function and the process in which venom proteins are involved.
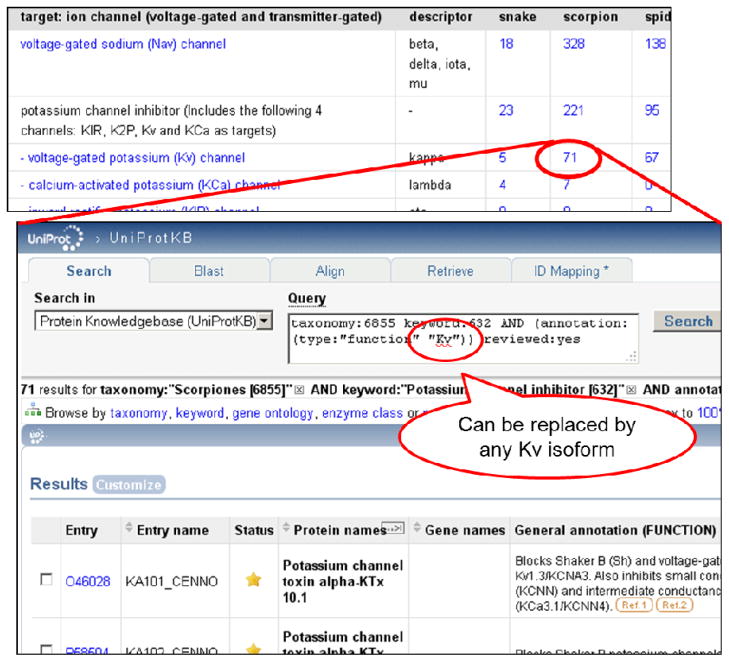
Information on venom proteins molecular target is provided in a table that lists the toxins targeting the different channel/receptor families according to the venomous phyla (http://www.uniprot.org/program/toxins/statistics). This table indicates the number of toxins of interest and provides links to retrieve them all. In addition, the queries are easy to build and modify (Fig. 2).
Figure 2.
Requests are book marked and the query is easy to edit and refine. For example, to retrieve scorpion toxins that target the subtype Kv1.1/KCNA1 of voltage-gated potassium channel, one should click on the case scorpion-Kv and replace ‘Kv’ by ‘Kv1.1’.
Many venom proteins display enzymatic activity, particularly snake toxins that often act as phospholipases, proteases, L-amino oxidases, and acetylcholinesterases. For such proteins, the Enzyme Commission (EC) number is indicated in ‘Names and origin’ and supplementary information is provided in the ‘General annotation’ field. This includes the ‘catalytic activity’ field used to describe the reaction(s) catalyzed by the enzyme, the ‘cofactor’ field that describes the non-protein substances required by the enzyme to be active, as well as the ‘enzyme regulation’ field that indicates regulatory mechanisms (e.g. UniProt ID: P0CB14). In addition, molecular function of enzymatic proteins is given in the topic ‘Function’, followed by its biological activity.
2.2.2 Post-translational modifications (PTMs)
Because protein toxins are often short and easy to isolate from entire venom, they are for the most part studied at protein level. Such studies have allowed extensive analysis of post-translational modifications (PTMs) of diverse chemical natures. These range from proteolytic cleavage and disulfide bonding to the addition of simple and complex chemical groups, such as amidation, hydroxylation, bromination, carboxylation, sulfation, N- or O-glycosylation, palmitoylation or N-terminal pyrrolidone carboxylic acid formation. D-amino acid isomerization is also found in animal toxins, but is rare. Although the precise function of the modified amino acids is generally unknown these modifications potentially contribute to variations in activity, structure and biological stability. Because of their biological importance and their utility as training data for predictive bioinformatics tools, UniProtKB/Swiss-Prot prioritizes the annotation of such PTMs (Farriol-Mathis et al., 2004), using data from the scientific literature and from protein 3D-structures.
PTMs are presented in UniProtKB/Swiss-Prot in a user-friendly, simple, and computer-readable manner. General information about PTMs is available under the topic ‘General annotation’, and most of these modifications are associated with keywords. The exact position of the modification is shown in the ‘Sequence annotation’ field (e.g. UniProt ID: P69770), while its chemical nature is indicated using strictly standardized annotation and controlled vocabularies (see www.uniprot.org/docs/ptmlist) based on the vocabulary provided by the RESID database (Garavelli, 2004). This database contains a comprehensive collection of PTMs with systematic and alternative names, formulas, and structure diagrams, and is linked to the PSI-MOD ontology useful in MS experiments, since it provides hierarchical representation of PTMs and serves as a tool for precisely annotating ambiguous or incomplete experimental results (Montecchi-Palazzi et al., 2008).
The accurate and structured format used by UniProtKB has several advantages for the community. Correctly assigned PTMs facilitate the correct identification of modified peptides by mass spectrometry. The structured format of the ‘Annotation sequence‘ field (i.e. ‘Chain’ or ‘Peptide’, ‘Sequence conflict’, ‘Natural variant’ and ‘Alternative sequence’) permits programmatic extraction and reconstruction of the different sequences that are merged in one entry and the computation of the theoretical peptide mass including the PTMs. In the knowledgebase, PTMs are propagated to closely related proteins. Such propagated PTMs are tagged with non-experimental qualifiers: ‘By similarity’ means that there is experimental evidence for a closely related protein; ‘Potential’ indicates data derived from the use of bioinformatics tools, e.g., for the prediction of cleavable signal sequences.
2.3 Nomenclature
Although the use of stable common names would facilitate the sharing of ideas and the finding of pertinent information there is not yet a unified animal toxin nomenclature.
Currently three nomenclature systems co-exist, for cone snail, for spider and for a class of scorpion toxins. The cone snail nomenclature proposed by Olivera and others (for a review, see McIntosh et al., 1999) is based on toxin pharmacological activity, cysteine scaffold and species of origin. This system is complex and is not always followed by the scientific community. An up-to-date description of this nomenclature is available in the ConoServer documents (http://www.conoserver.org/?page=about_conotoxins&bpage=cononames) (Kaas et al., 2012). The spider nomenclature is based on pharmacological activity, taxonomy, and species of origin. This nomenclature has been recently proposed by King et al. (2008), and all spider toxins have been renamed accordingly in ArachnoServer. Sea anemone toxins are also classified based on the spider nomenclature scheme, and on the cysteine scaffold (Kozlov and Grishin, 2012). The last nomenclature system concerns the scorpion toxins that specifically target potassium channels. It is based on amino acid sequence motifs and on the location of cysteine residues that are crucial for 3D-structure (de la Vega and Possani, 2004; Tytgat et al., 1999). For this specific case, UniProtKB provides a document, called ‘scorpktx.txt’, that lists all toxins known to date, according to this nomenclature system and their mapping to the knowledgebase (http://www.uniprot.org/docs/scorpktx). References to these different nomenclature systems can be retrieved at http://www.uniprot.org/docs/nomlist.
In UniProtKB/Swiss-Prot, all protein names and gene names where available, are stored in ‘Names and origin’ (e.g. UniProt ID: P01495). Toxin names that follow established nomenclature guidelines are stored as ‘Recommended name’, i.e. names that we would suggest to use when citing proteins. For names not subjected to any nomenclature system the recommended name is left to curator judgment. Whenever possible, the most frequently used name is chosen. In all cases, synonyms found in the literature or in other resources, such as organism specific databases or nucleotide sequence database, are kept as ‘Alternative name(s)’.
2.4 Taxonomy
Each UniProtKB entry indicates the scientific name of the source organism of the protein concerned, in addition to the English common name and a synonym when available, the taxonomic lineage, the taxonomic identifier (TaxID) which is provided by the NCBI (e.g. UniProt ID: P56676), as well as the Organism Species code (OS code), i.e. the five-letter mnemonic code which appears in the protein entry identifier of the knowledgebase (e.g. MESMA that stands for MESobuthus MArtensii). To provide these different data, UniProt maintains a database of taxonomy (http://www.uniprot.org/taxonomy/) (Phan et al., 2003) to which new species can be submitted during the curation process (Fig. 1). Each organism in this database has a unique taxonomic identifier (TaxID) which is provided by the NCBI. If no TaxId is already assigned to the organism, the species is submitted to the NCBI, which attributes a new one. In all cases, the lineage is checked and a common name as well as a synonym, when available, is added. Detection of classification problems allows corrections in the NCBI, which, as shown in Fig. 1, improves the level of correctness in the three databases (taxonomy of NCBI, INSDC and UniProt).
2.5 Database cross-references
Among the over 120 cross-references provided by UniProtKB some are of particular relevance for the toxin field. Obviously both ConoServer and ArachnoServer databases are cross-linked with UniProtKB/Swiss-Prot entries.
2.5.1 Links to nucleotide sequence databases
UniProtKB provides links to the INSDC, where available. For some toxins the protein sequences were translated from nucleotide sequences submitted to INSDC without indication of the coding sequence (CDS). These will not appear in UniProtKB/TrEMBL. They might, however, be present in UniProtKB/Swiss-Prot and will be associated with the tag ‘No translation available’ indicated in the cross-reference section.
2.5.2 Links to 3D-structure databases
Due to their pharmacological importance, toxin 3D-structures are extensively studied and those that are submitted to the Protein Data Bank (PDB) (Markley et al., 2008) can be retrieved through cross-references from UniProtKB. Currently, about 8% of the venom proteins annotated in UniProtKB/Swiss-Prot have an experimentally determined 3D-structure, while the overall proportion in the knowledgebase is inferior to 0.1% (UniProt release 2012_2). Visualization tools to apprehend the structure are also provided in UniProtKB through a cross-reference to PDBsum (Laskowski, 2009). This database summarizes information about each experimentally determined structure in PDB, and provides tools to visualize 3D-structures or to load structures, such as AstexViewer (Hartshorn, 2002), for example. About half the venom proteins contain a cross-reference to the Protein Model Portal (PMP) (Arnold et al., 2009) that gives access to various models, and provides access to interactive services for model building, and quality assessment. In addition to the cross-reference to 3D-structure databases, information inferred from 3D-structure is indicated, for example, key residues that are involved in catalysis, or that bind metal ions as well as cysteines disulfide-bonded in the ‘Sequence annotation’ topic (e.g. UniProt ID: Q6SLM1).
2.5.3 Links to family and domain databases
Domains and repeats are the basic building blocks of proteins, and the combination of several such modules contributes to the evolution of functional diversity in proteins (Moore et al., 2008). These are catalogued in the InterPro database (Hunter et al., 2009), which integrates predictive models or ‘signatures’ representing protein domains and families from diverse member databases. The InterPro member databases are cross-referenced in the ‘Family and domain databases’ category. However, these databases do not provide high coverage yet for short toxins, such as conotoxins, and only a few of them, if any, are cross-referenced from these venom protein entries. In contrast, most long toxins (such as snake toxins for example) belong to well-studied protein families, most probably because they were recruited into the venom proteomes from ‘old’ protein families during evolution (Fry, 2005). In addition to the cross-reference to ‘Family and domain databases’, UniProtKB/Swiss-Prot indicates the presence of particular domains or repeats in the ‘General annotation’ field under the topic ‘Sequence similarities’. The exact extent of such domains, repeats, and sequence motifs are displayed in the ‘Sequence annotation’ section. It is worthy to note, however, that membership of a protein family is not an infallible indicator of protein function. As an example, the snake phospholipase A2 (UniProt ID: Q9IAT9) belongs to the phospholipase A2 family but lacks this enzymatic activity.
3. UniProt website
The UniProt website www.uniprot.org (Jain et al., 2009) allows the easy retrieval of protein(s) of interest through the use of different tools such as full text and field-based search, sequence similarity search, multiple sequence alignment, batch retrieval and database identifier mapping. Most data (including documentation and help) can be searched through the full text search, which allows searches requiring no prior knowledge of our data or search syntax. Results are sorted by relevance and, suggestions are provided to help refine searches that yield too many or no results. The field-based text search supports more complex queries. These can be built iteratively with the tool bar query builder or entered manually in the query field. In addition, the site has a simple and consistent URL scheme and all searches can be book marked to be repeated at a later time.
To simplify retrieval of toxins of interest, different tables have been created and can be displayed on the UniProt website:
Toxins or venom proteins that are secreted by the different venomous/poisonous phyla (see the first table of http://www.uniprot.org/program/toxins/statistics).
Toxins targeting the different channel/receptor families in function of the different venomous phyla (see also paragraph 2.2.1 for details and second table of http://www.uniprot.org/program/toxins/statistics).
Scorpion potassium channel toxins (see also paragraph 2.3 for details and http://www.uniprot.org/docs/scorpktx).
Statistics on venom proteins such as ‘Cross-references to PDB’ and ‘PubMed citations’ in snakes, cone snails, scorpions and spiders (http://www.uniprot.org/program/toxins/statistics).
4. Submission of new protein sequences
Similarly to the need for a rapid access to detailed information about venom proteins, a rapid access to publicly available sequences is a necessity. Up to now, nucleotide sequences have been regularly submitted to the INSDC, thanks to publishers who requested an accession number to include in publications. In a similar manner, sequences obtained by direct protein sequencing (only Edman degradation and MS/MS de novo sequencing) and experimental results can be submitted to UniProtKB. This can be done via the SPIN tool (http://www.ebi.ac.uk/swissprot/Submissions/spin/). The authors and the scientific community will benefit from such submissions. Authors will have an accession number to cite in their publication and their sequences will be available both in UniProtKB/Swiss-Prot and in the GenPept section of NCBI, since the NCBI regularly imports UniProtKB/Swiss-Prot entries. Such a submission will also enhance dissemination of the identified venom protein and associated data. In addition, the submission will allow linking of disparate information together (function, organism, publication/submitter, or PTMs) even if the sequence is only a fragment, and it will permit a rapid overview of what is known on venom composition.
Sequences submitted to UniProtKB can be kept confidential for a reasonable time (until publication, for example). In addition to sequence, and associated protein name, organism and submitter’s name/reference, more information is highly welcome. For batch submission, help@uniprot.org should be contacted.
5. Current status
UniProtKB/Swiss-Prot currently contains 5’090 venom protein entries from 489 venomous species (UniProt release 2012_02). For more detailed statistics, see the statistic page cited above.
UniProtKB/TrEMBL currently contains 2’249 proteins entries that are potentially coming from venom. They can be retrieved thanks to this link: http://doiop.com/r1b5f3
6. Conclusions
UniProtKB/Swiss-Prot is a freely accessible resource that offers accurate and concise information about toxin proteins produced by venomous animals through its animal toxin annotation program. This information includes toxin sequences, functional annotation, structural information, PTMs, and information on toxin targets, which are also fully annotated. Through careful literature curation and the incorporation of direct sequence submissions from authors, UniProtKB/Swiss-Prot provides access to toxin sequences known only at the protein sequence level, and which would otherwise be unavailable to the scientific community. UniProtKB/Swiss-Prot contains toxins from all venomous phyla, a feature that facilitates comparative studies of toxin evolution and action. UniProtKB/Swiss-Prot complements other specialized resources providing detailed information on toxins from spiders and cone snails, and serves as the major reference for toxins from snakes, scorpions, and sea anenomes, which are not served by specialist databases. These will remain a clear priority for our future efforts.
Acknowledgments
The authors would like to thank Alan Bridge for critical reading and correction of the manuscript. The Swiss-Prot group is part of the Swiss Institute of Bioinformatics (SIB) and of the UniProt consortium. Its activities are supported by the Swiss Federal Government through the Federal Office of Education and Science and by the National Institutes of Health (NIH) grant 1 U41 HG006104-01. Additional support comes from the European Commission contract SLING (226073).
Abbreviations
- HGNC
HUGO Gene Nomenclature Committee
- INSDC
International Nucleotide Sequence Database Collaboration
- IUPHAR
International Union of Basic and Clinical Pharmacology nomenclature
- MS
mass spectrometry
- PTM
post-translational modification
Footnotes
Conflicts of interest
None to declare.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alexander S, Harmar A, McGrath I. New updated GRAC Fifth Edition with searchable online version Launch of new portal Guide to Pharmacology in association with NC-IUPHAR Transporter-Themed Issue. Br J Pharmacol. 2011;164:1749–1750. doi: 10.1111/j.1476-5381.2011.01751.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold K, Kiefer F, Kopp J, Battey JN, Podvinec M, Westbrook JD, Berman HM, Bordoli L, Schwede T. The protein Model Portal (PMP) J Struct Funct Genomics. 2009;10:1–8. doi: 10.1007/s10969-008-9048-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohlen CJ, Chesler AT, Sharif-Naeini R, Medzihradszky KF, Zhou S, King D, Sanchez EE, Burlingame AL, Basbaum AI, Julius D. A heteromeric Texas coral snake toxin targets acid-sensing ion channels to produce pain. Nature. 2011;479:410–414. doi: 10.1038/nature10607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Vega RRC, Possani LD. Current views on scorpion toxins specific for K+-channels. Toxicon. 2004;43:865–875. doi: 10.1016/j.toxicon.2004.03.022. [DOI] [PubMed] [Google Scholar]
- Escoubas P, Sollod B, King GF. Venom landscapes: mining the complexity of spider venoms via a combined cDNA and mass spectrometric approach. Toxicon. 2006;47:650–663. doi: 10.1016/j.toxicon.2006.01.018. [DOI] [PubMed] [Google Scholar]
- Farriol-Mathis N, Garavelli JS, Boeckmann B, Duvaud S, Gasteiger E, Gateau A, Veuthey A, Bairoch A. Annotation of post-translational modifications in the Swiss-Prot knowledgebase. Proteomics. 2004;4:1537–1550. doi: 10.1002/pmic.200300764. [DOI] [PubMed] [Google Scholar]
- Fry BG. From genome to “venome”: molecular origin and evolution of the snake venom proteome inferred from phylogenetic analysis of toxin sequences and related body proteins. Genome Res. 2005;15:403–420. doi: 10.1101/gr.3228405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garavelli JS. The RESID Database of Protein Modifications as a resource and annotation tool. Proteomics. 2004;4:1527–1533. doi: 10.1002/pmic.200300777. [DOI] [PubMed] [Google Scholar]
- Gowd KH, Dewan KK, Iengar P, Krishnan KS, Balaram P. Probing peptide libraries from Conus achatinus using mass spectrometry and cDNA sequencing: identification of δ and ω-conotoxins. J Mass Spectrom. 2008;10:141–155. doi: 10.1002/jms.1377. [DOI] [PubMed] [Google Scholar]
- Hartshorn MJ. AstexViewer: A visualisation aid for structure-based drug design. J Comput Aided Mol Des. 2002;16:871–881. doi: 10.1023/a:1023813504011. [DOI] [PubMed] [Google Scholar]
- He Q, Han W, He Q, Huo L, Zhang J, Lin Y, Chen P, Liang S. ATDB 2.0: A database integrated toxin-ion channel interaction data. Toxicon. 2010;56:644–647. doi: 10.1016/j.toxicon.2010.05.013. [DOI] [PubMed] [Google Scholar]
- Herzig V, Wood DLA, Newell F, Chaumeil P-C, Kaas Q, Binford GJ, Nicholson GM, Gorse D, King GF. ArachnoServer 2.0, an updated online resource for spider toxin sequences and structures. Nucleic Acids Res. 2010;39:653–657. doi: 10.1093/nar/gkq1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter S, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, Bork P, Das U, Daugherty L, Duquenne L, Finn RD, Gough J, Haft D, Hulo N, Kahn D, Kelly E, Laugraud A, Letunic I, Lonsdale D, Lopezm R, Madera M, Maslen J, McAnulla C, McDowall J, Mistry J, Mitchell A, Mulder N, Natale D, Orengo C, Quinn AF, Selengut JD, Sigrist CJ, Thimma M, Thomas PD, Valentin F, Wilson D, Wu CH, Yeats C. InterPro: the integrative protein signature database. Nucleic Acids Res. 2009;37:D211–D215. doi: 10.1093/nar/gkn785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain E, Bairoch A, Duvaud S, Phan I, Redaschi N, Suzek BE, Martin MJ, McGarvey P, Gasteiger E. Infrastructure for the life sciences: design and implementation of the UniProt website. BMC Bioinformatics. 2009;10:136–155. doi: 10.1186/1471-2105-10-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakubowski JA, Keays DA, Kelley WP, Sandall DW, Bingham JP, Livett BG, Gayler KR, Sweedler JV. Determining sequences and post-translational modifications of novel conotoxins in Conus victoriae using cDNA sequencing and mass spectrometry. J Mass Spectrom. 2004;39:548–557. doi: 10.1002/jms.624. [DOI] [PubMed] [Google Scholar]
- Jungo F, Bairoch A. Tox-Prot, the toxin protein annotation program of the Swiss-Prot protein knowledgebase. Toxicon. 2005;45:293–301. doi: 10.1016/j.toxicon.2004.10.018. [DOI] [PubMed] [Google Scholar]
- Jungo F, Estreicher A, Bairoch A, Bougueleret L, Xenarios I. Animal Toxins: How is Complexity Represented in Databases? Toxin. 2010;2:262–282. doi: 10.3390/toxins2020262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaas Q, Yu R, Jin AH, Dutertre S, Craik DJ. ConoServer: updated content, knowledge, and discovery tools in the conopeptide database. Nucleic Acids Res. 2012;40:D325–D330. doi: 10.1093/nar/gkr886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karsch-Mizrachi I, Nakamura Y, Cochrane G. On behalf of the International Nucleotide Sequence Database Collaboration. The International Nucleotide Sequence Database Collaboration. Nucleic Acids Res. 2012;40:D33–D37. doi: 10.1093/nar/gkr1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King GF, Gentz MC, Escoubas P, Nicholson GM. A rational nomenclature for naming peptide toxins from spiders and other venomous animals. Toxicon. 2008;52:264–276. doi: 10.1016/j.toxicon.2008.05.020. [DOI] [PubMed] [Google Scholar]
- Koh CY, Kini RM. From snake venom toxins to therapeutics - Cardiovascular examples. Toxicon. 2012;59:497–506. doi: 10.1016/j.toxicon.2011.03.017. [DOI] [PubMed] [Google Scholar]
- Kordis D, Gubensek F. Adaptive evolution of animal toxin multigene families. Gene. 2000;261:43–52. doi: 10.1016/s0378-1119(00)00490-x. [DOI] [PubMed] [Google Scholar]
- Kozlov S, Grishin E. Convenient nomenclature of cysteine-rich polypeptide toxins from sea anemones. Peptides. 2012 doi: 10.1016/j.peptides.2011.12.008. In press. [DOI] [PubMed] [Google Scholar]
- Laskowski RA. PDBsum new things. Nucleic Acids Res. 2009;37:D355–D359. doi: 10.1093/nar/gkn860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magrane M UniProt Consortium. UniProt Knowledgebase: a hub of integrated protein data. Database. 2011;29:bar009. doi: 10.1093/database/bar009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markley JL, Ulrich EL, Berman HM, Henrick K, Nakamura H, Akutsu H. BioMagResBank (BMRB) as a partner in the Worldwide Protein Data Bank (wwPDB): New policies affecting biomolecular NMR depositions. J Biomol NMR. 2008;40:153–155. doi: 10.1007/s10858-008-9221-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh JM, Santos AD, Olivera BM. Conus peptides targeted to specific nicotinic acetylcholine receptor subtypes. Annu Rev Biochem. 1999;68:59–88. doi: 10.1146/annurev.biochem.68.1.59. [DOI] [PubMed] [Google Scholar]
- Montecchi-Palazzi L, Beavis R, Binz PA, Chalkley RJ, Cottrell J, Creasy D, Shofstahl J, Seymour SL, Garavelli JS. The PSI-MOD community standard for representation of protein modification data. Nat Biotechnol. 2008;26:864–866. doi: 10.1038/nbt0808-864. [DOI] [PubMed] [Google Scholar]
- Moore AD, Björklund AK, Ekman D, Bornberg-Bauer E, Elofsson A. Arrangements in the modular evolution of proteins. Trends Biochem Sci. 2008;33:444–451. doi: 10.1016/j.tibs.2008.05.008. [DOI] [PubMed] [Google Scholar]
- Phan IQ, Pilbout SF, Fleischmann W, Bairoch A. NEWT, a new taxonomy portal. Nucleic Acids Res. 2003;31:3822–3823. doi: 10.1093/nar/gkg516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seal RL, Gordon SM, Lush MJ, Wright MW, Bruford EA. Genenames.org: the HGNC resources in 2011. Nucleic Acids Res. 2011;39:D514–D519. doi: 10.1093/nar/gkq892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The UniProt Consortium. Reorganizing the protein space at the Universal Protein Resource (UniProt) Nucleic Acids Res. 2012;40:D71–D75. doi: 10.1093/nar/gkr981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tytgat J, Chandy KG, Garcia ML, Gutman GA, Martin-Eauclaire MF, van der Walt JJ, Possani LD. A unified nomenclature for short-chain peptides isolated from scorpion venoms: alpha-KTx molecular subfamilies. Trends Pharmacol Sci. 1999;20:444–447. doi: 10.1016/s0165-6147(99)01398-x. [DOI] [PubMed] [Google Scholar]