Abstract
We investigated the genetic causes of ethanol tolerance by characterizing mutations selected in Saccharomyces cerevisiae W303-1A under the selective pressure of ethanol. W303-1A was subjected to three rounds of turbidostat, in medium supplemented with increasing amounts of ethanol. By the end of selection, the growth rate of the culture has increased from 0.029 h-1 to 0.32 h-1. Unlike the progenitor strain, all yeast cells isolated from this population were able to form colonies on medium supplemented with 7% ethanol within six days, our definition of ethanol tolerance. Several clones selected from all three stages of selection were able to form dense colonies within two days on solid medium supplemented with 9% ethanol. We sequenced the whole genomes of 6 clones and identified mutations responsible for ethanol tolerance. Thirteen additional clones were tested for the presence of similar mutations. In 15 out of 19 tolerant clones the stop-codon in ssd1-d was replaced with an aminoacid-encoding codon. Three other clones contained one of two mutations in UTH1, and one clone did not contain mutations in either SSD1 or UTH1. We showed that the mutations in SSD1 and UTH1 increased tolerance of the cell wall to zymolyase and conclude that stability of the cell wall is a major factor in increased tolerance to ethanol.
Keywords: ethanol tolerance, SSD1, UTH1, turbidostat, cell wall
Introduction
In addition to being a model organism for studying eukaryotic biology, yeast has become central in microbiological research due to its ability to produce ethanol. It has been used for brewing during most of human history in all civilizations, and recent interest in large-scale biofuel industry (Sanchez & Cardona, 2008) has reinvigorated interest in the biological basis of ethanol production. Saccharomyces cerevisiae remains a vital alternative to numerous other fermentative microorganisms (Lin & Tanaka, 2006) as an industrial ethanol producer. High-titer fermentation requires elevated resistance of yeast to inhibitory ethanol concentrations and thus elucidation of the genetic and physiological underpinnings of ethanol tolerance in S. cerevisiae is an important theoretical as well as practical challenge (for recent reviews see Ding, et al., 2009, Yoshikawa, et al., 2009, Ma & Liu, 2010).
Ethanol tolerance is usually considered to be the ability of yeast to survive short-time exposure to elevated ethanol concentrations (Kim, et al., 1996, Sharma, 1997, Yamaji, et al., 2003). Survival under high ethanol concentrations requires activation of the general stress response (Ogawa, et al., 2000, Yamaji, et al., 2003) and probably other, more specific, pathways (Alexandre, et al., 2001). In this work we studied the genetic basis of ethanol tolerance by selecting mutations that affected the proliferation rate of yeast in the presence of ethanol, rather than those that affected survival alone. It is known that yeast strains differ widely in their ability to proliferate in the presence of ethanol. For example, yeast used for Sake production forms colonies on solid medium containing 8-9% ethanol within a few days, while the laboratory strain W303-1A does not do so after a week in the presence of 6% ethanol (this work). The ability to form colonies on solid medium containing at least 7% ethanol within 6 days was used here as the operative definition of ethanol tolerance. Thus, Sake yeast was defined as ethanol-tolerant while W303-1A is ethanol-sensitive. Sake yeasts are so different from common S. cerevisiae strains in their regulation of gene expression and even in their karyotype (Akao, et al., 2011) that it is practically impossible to relate with certainty any single genomic difference to their ethanol tolerance phenotype, without extensive quantitative trait locus (QTL) analysis. However, since Sake yeast is also not readily accessible to genetic manipulations (Nakazawa, 1993), such QTL analysis is not feasible. Thus, for these reasons, and because we set out to identify mutations that could be selected as a result of exposure to high concentrations of ethanol, it was imperative that we begin with a sensitive strain, rather than one already capable of growing at high ethanol concentrations. We thus used W303-1A for our experiments.
Many different approaches have been used to study the molecular basis of ethanol tolerance. Deletion libraries of laboratory yeast have been used to identify genes whose deletions result in ethanol-sensitivity (Takahashi, et al., 2001, Kubota, et al., 2004, Fujita, et al., 2006, van Voorst, et al., 2006). These studies have generated long lists of genes belonging to almost every physiological pathway.
A further strategy has been to identify genes, whose transcription is up- or down-regulated in response to ethanol (Ogawa, et al., 2000, Alexandre, et al., 2001, Chandler, et al., 2004, Hirasawa, et al., 2007). This approach has also revealed dozens of genes, though they are not necessarily directly related to ethanol tolerance, because in most cases, it has not been shown that their overexpression results in a tolerant phenotype. One exclusion to this rule is over-expression of the TRP1-5 genes (Hirasawa, et al., 2007) that does result in an increased growth rate.
A few studies have reported isolation from continuous culture of yeast more ethanol-tolerant than their parental strains (Brown & Oliver, 1982, Jimenez & Benitez, 1988, Hirasawa, et al., 2007, Yazawa, et al., 2007, Stanley, et al., 2010), but while the tolerant mutants were characterized physiologically, the genetic bases of their tolerance have not yet been explored.
The existing data provide little mechanistic understanding of the phenomenon of ethanol tolerance. It has been suggested that it is an integrative phenomenon requiring adaptations to occur in multiple metabolic pathways, involving changes in, among other things, cell wall and membrane stability and general stress tolerance (Ogawa, et al., 2000). Alternatively, it has been claimed that a small number of “master” genes (or, perhaps, even one) play a central role in ethanol tolerance (Alper, et al., 2006), though these genes have not yet been identified with certainty (Baerends, et al., 2009).
In this study, we wished to use an approach that is unbiased toward the underlying mechanisms or the specific genes. We therefore decided to select for an ethanol-tolerant population in continuous culture under the strict selection pressure of sub-lethal ethanol concentration. Recent advances in whole genome sequencing allowed for the elucidation of the selected genetic changes.
Continuous culture has long been an important tool in industrial strain development (Bloom & Mcfall, 1975, Harder, et al., 1977, Bennett & Boraas, 1988), and has even been used to study ethanol tolerance in yeast. For example, Brown & Oliver selected ethanol-tolerant S. uvarum strains in a chemostat by steadily increasing the ethanol concentration in proportion to the CO2 output of the population (Brown & Oliver, 1982). In the course of their experiment, the concentration of ethanol in the medium more than doubled (from 2% to 4.7%), and several clones were isolated with improved fermentation capacity in 10% ethanol. Similarly, Jimenez and Benitez selected for ethanol-tolerant hybrids between industrial wine strains and laboratory strains, by increasing the ethanol concentration in the culture in response to reductions in pH (Jimenez & Benitez, 1988). Most recently, Stanley et al. used directed evolution, under the selection pressure of sub-lethal ethanol concentrations (Stanley, et al., 2010). The chemostat was operated at a constant dilution rate, and the ethanol concentration was increased when the growth rate had become equal to the dilution rate. This system allowed for the selection of spontaneous and chemically-induced mutations in S. cerevisiae W303-1A that resulted in increased acclimation and growth rates when cultivated at sub-lethal (6%) ethanol concentration. These strains also showed increased viability at normally lethal concentrations of ethanol (Stanley, et al., 2010). The selection required continuous operation for 192 days with the un-mutated population, and 14 to 28 days following treatment with a chemical mutagen. The genetic basis for the increased tolerance was not elucidated.
In this work, we modified the strategy of Stanley et al, and imposed the selection pressure on a cell population grown in a turbidostat, i.e., a system in which the population is maintained at a constant biomass concentration (Watson, 1972). The rationale was to give a large population of cells the time and opportunity to adapt (via accumulating genetic or epigenetic changes). Given the known mutation rate (Lang & Murray, 2008) and the population size, we would expect there to be clonal interference (Kao & Sherlock, 2008), and thus the existence of a mixed population (Kopp & Hermisson, 2009), composed of several sub-populations, possibly acquiring ethanol tolerance by different mechanisms, with different genes involved.
We subjected the laboratory yeast strain W303-1A to selection in a turbidostat under increasing ethanol concentrations. In a relatively short time (3 weeks - 141 generations compared to 23 weeks and 486 generations in (Stanley, et al., 2010)), we obtained a population whose proliferation rate was an order of magnitude (0.029 h-1 versus 0.32 h-1) better in the presence of 8% ethanol compared to the parental strain. Analysis of the selected population showed that it is heterogeneous with respect to its capabilities of growth in the presence of ethanol. We isolated and analyzed a total of 6 sub-clones of this population (which grew best in the presence of ethanol) via whole genome sequencing.
We report that in each of these clones a single point mutation either in SSD1 or UTH1 was sufficient to improve the growth rate in the presence of ethanol.
Materials and Methods
Yeast strains, media and general methods
The S. cerevisiae strains used in this study are listed in Table 1. All experiments were performed with isogenic strains of W303-1A. The uth1Δ mutant was created by disrupting the UTH1 open reading frame with the gentamicin tolerance gene KanMX4 using a PCR-based knockout strategy (Brachmann, et al., 1998) and verified by PCR. Strains were grown at 30°C on YPD medium containing (gL-1) yeast extract 10, Bactopeptone 20 and glucose, 20, or on the synthetic medium YNB-Ura containing (gL-1) yeast nitrogen base 1.7, ammonium sulfate 5, glucose 20, leucine, adenine, tryptophane, uracil, histidine, lysine and methionine 0.04. Solid media were made with 3% agar. Strains were grown in liquid or solid media supplemented with ethanol as indicated in each experiment. Petri dishes with ethanol-containing solid media were sealed with parafilm to avoid evaporation.
Table 1.
Yeast strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| W303-1A | MATa mal can1-100 ade2-1 his3-11,15 leu2-3,13 trp1-1 ura3-1 ssd1-d | Yeast genetic Stock Center, Berkeley, CA |
| W303-1Aα | MATα mal can1-100 ade2-1 his3-11,15 leu2-3,13 trp1-1 ura3-1 ssd1-d | This work |
| W303-1A uth1Δ | MATa mal can1-100 ade2-1 his3-11,15 leu2-3,13 trp1-1 ura3-1 uth1∷kanMX4 | This work |
| Kyokai 9 (K9) Sake yeast | Haruyuki Iefugi (Shobayashi, et al., 2007) |
Yeast was transformed using the lithium acetate method (Gietz & Schiestl, 2007). Genetic crosses of haploid strains, sporulation of diploid strains, and tetrad analysis were carried out as described by (Sherman & Hicks, 1991).
Turbidostat selection procedure
W303-1A was grown in Erlenmeyer flasks (250 ml) containing 50ml YPD for 18 hours in a rotary shaker (250 rpm, 2 inch, 30°C). Cells were pelleted by centrifuging, re-suspended in the same volume (50ml) of YPD medium supplemented with 6% (vol/vol) ethanol and returned to the shaker for an additional 24 hours. This culture was used as the inoculum for the turbidostat. A 250ml turbidostat vessel with a working volume of 120 ml, aerated at 0.1 vvm, was inoculated at OD600 = 1 in YPD supplemented with 6% (vol/vol) ethanol. According to optical density, the initial number of cells in the vessel was, ca., 4×1010. The growth rate, μ, and the dilution rate, D, were monitored once per day. D was calculated as:
where Veffl is the volume of the effluent, Vferm is the fermentation volume and t is the fermentation time in hours. The growth rate was calculated as:
where Nt and N0 are culture densities at the end of measured fermentation period and at the beginning of it, respectively. The dilution rate was manually adjusted to the same value as the growth rate.
The turbidostat was operated at 30°C for 8 days (Stage I); the culture was then harvested and the half of it was used as inoculum for Stage II. The Stage II turbidostat culture was resumed for the additional 7 days as described above but in YPD supplemented with 7% (vol/vol) of ethanol. At the end of 7 days, the culture was supplemented with ethanol to the final concentration of 8% (vol/vol). This Stage III culture was grown for another 7 days.
Determination of maximal growth rate (μmax)
The strain W303-1A, as well as clones 9C and 9E were grown overnight in New Brunswick Renova rotatory shaker at 250 rpm in Erlenmeyer flasks (250 ml) containing YPD medium (50 ml) at 30°C. The cultures were then seeded in YPD medium supplemented with 8% ethanol at OD600 = 0.25. The growth was carried out at the same conditions as shown above. The cultures were sampled at intervals, which allowed collecting 6-10 readings of OD600 during the exponential phase of growth. The maximal growth rate (μmax) was calculated using at least four independent experiments.
Plasmid Construction
Plasmids and primers used in the study are listed in Tables 2 and 3 respectively.
Table 2.
Plasmids used in this study
| Plasmid | Description | Source |
|---|---|---|
| pRS416 | Yeast centromeric vector containing the CEN element and the URA3. | (Sikorski & Hieter, 1989) |
| pRS306 | Yeast integrative vector. | (Sikorski & Hieter, 1989) |
| pRS306-UTH1 D224Y | pRS306 containing the UTH1D224Y with 700 bp upstream and 400bp downstream. | This work |
| pRS306-UTH1WT | pRS306 containing the UTH1 with 700 bp upstream and 400bp downstream. | This work |
| pRS416-SSD1-V | pRS416 containing the SSD1-V with 399 bp upstream and 404bp downstream. | (Reinke, et al., 2004) |
| pRS416-ssd1-d | pRS416 containing the ssd1-d with 399 bp upstream and 404bp downstream. | (Reinke, et al., 2004) |
| pRS416-ssd1-dstop698W | pRS416, containing the ssd1-dstop698W with 399 bp upstream and 404bp downstream. | This work |
| pRS416-ssd1-dstop698S | pRS416, containing the ssd1-dstop698S with 399 bp upstream and 404bp downstream. | This work |
| pRS416-ssd1-dstop698E | pRS416, containing the ssd1-dstop698E with 399 bp upstream and 404bp downstream. | This work |
| pRS416-ssd1-dstop698Q | pRS416, containing the ssd1-dstop698Q with 399 bp upstream and 404bp downstream. | This work |
Table 3.
Primers used in this study
| Primer name | Used for | Primer sequence (5’ to 3’) |
|---|---|---|
| UTH1_F | Sequence from inside UTH1 toward position 224 | GTCCACCATAGTGACAACCAC |
| UTH1_R | Sequence from inside UTH1 toward position 224 | AGCAAGCACCATTGACGGTAG |
| SSD1_736_F | Sequence from inside ssd1-d toward position 224 | GACGATGAATTCATAGCAACCTCTTC |
| SSD1_2296_ R | Sequence from inside SSD1 toward position 224 | CGGGATCCGTTCGTCGTTGAACGATTG |
| ssd1-d-stop698W-F | Mutation of ssd1-d at stop 698 to Trp | CGGACACTAATGAGTGGAATATCTTTGCAATTTCCGAGC |
| ssd1-d-stop698W-R | Mutation of ssd1-d at stop 698 to Trp | GCTCGGAAATTGCAAAGATATTCCACTCATTAGTGTCCG |
| ssd1-d-stop698S- F | Mutation of ssd1-d at stop 698 to Cys | CGGACACTAATGAGTCGAATATCTTTGCAATTTCCGAGC |
| ssd1-d-stop698S- R | Mutation of ssd1-d at stop 698 to Cys | GCTCGGAAATTGCAAAGATATTCGACTCATTAGTGTCCG |
| ssd1-d-stop698E- F | Mutation of ssd1-d at stop 698 to Glu | CGGACACTAATGAGGAGAATATCTTTGCAATTTCCGAGC |
| ssd1-d-stop698E- R | Mutation of ssd1-d at stop 698 to Glu | GCTCGGAAATTGCAAAGATATTCTCCTCATTAGTGTCCG |
| ssd1-d-stop698Q- F | Mutation of ssd1-d at stop 698 to Gln | CGGACACTAATGAGCAGAATATCTTTGCAATTTCCGAGC |
| ssd1-d-stop698Q- R | Mutation of ssd1-d at stop 698 to Gln | GCTCGGAAATTGCAAAGATATTCTGCTCATTAGTGTCCG |
| UTH1_EcoRI_F | Clone UTH1 gene from W303-1A, 9C or BY4741 uht1∷ KanMX4 (for knockout) | GCCGGAATTCCACCCGGACAAACATCGTTATC (EcoRI restriction site is shown in bold and underlined). |
| UTH1_BamHI_R | Clone UTH1 gene from W303-1A, 9C or BY4741 uht1∷ KanMX4 (for knockout) | CCGCGGATCCCATTTGTCTCACCACCAGAG (BamHI restriction site is shown in bold and underlined). |
Forward primers are indicated by ‘F’, reverse primers by ‘R’.
The SSD1-V allele was isolated from plasmid pPL094 (a kind gift from Ted Power, (Reinke, et al., 2004)), using the restriction enzymes XhoI and SacI, and ligated into pRS416 in order to obtain the pRS416-SSD1-V plasmid. The ssd1-d allele was extracted from pPL093 (a kind gift from Ted Power, (Reinke, et al., 2004)) using the restriction enzymes SpeI and XhoI and ligated into pRS416 in order to obtain the pRS416-ssd1-d plasmid. Each of the SSD1 alleles, either SSD1-V or ssd1-d, in the constructs contain 399 bp upstream and 404 bp downstream of the ORF.
UTH1 variant alleles were amplified from genomic DNA of strains W303-1A or 9C, using colony PCR. The UTH1 coding region, including 700 bp upstream and 400 bp downstream, was amplified with the forward primer containing an EcoRI restriction site and the reverse primer containing a BamHI restriction. The PCR products were introduced by ligation into the vector pRS306 that had been digested with EcoRI and BamHI using standard recombinant DNA techniques.
Site directed Mutagenesis
Site directed mutagenesis was carried out using QuickChange® kit (STRATAGENE). It was performed according to manufacturer’s instructions using relevant primers and pRS416-ssd1-d as the template.
Spot assay and growth condition
Yeast cells were cultured overnight in liquid medium (YNB-Ura or YPD). The cells were diluted in the appropriate medium to OD600 =0.2, grown for 2-5 hours and harvested in early logarithmic phase (OD600 ≈0.4-0.6). The cells were re-suspended in a fresh growth medium to a concentration of 107 cells per mL. Serial dilutions of the cultures were made (107, 106, 105, 104 and 103 cells per mL). An aliquot of each dilution (5μl) was deposited on an indicated 3% agar medium and cultured at 30°C for several days until small colonies developed. Alternatively, the dilution was made by streaking cultures of equal cell concentration on 3% agar; cell proliferation rate of various yeast isolates was compared by observation following several days in culture. All assays were performed with several concentrations of ethanol at least three times for each concentration; all assays were consistently reproducible.
Stress resistance
Oxidative stress was estimated by spotting yeast culture on YNB solid medium (agar, 3%, yeast nitrogen base, 0.17%, ammonium sulfate, 0.5%, glucose 2%) containing 1mM H2O2 and allowing it to grow for 48 hours at 30°C. Caffeine tolerance was estimated by spotting yeast on YPD agar containing 9 mM caffeine and assaying growth at 30°C.
Sensitivity of yeast cell wall to zymolyase
The cell wall lysis assay was performed as described in (Ovalle, et al., 1998). Briefly, cells were grown overnight in rich media at 30°C, harvested by centrifuging, washed three times with deionized water, and re-suspended at OD600 = 0.5 in TE buffer (50 mM Tris/HCl, 5 mM EDTA, pH 7.5). zymolyase 20T, 0.5U (20 U mg-1; Seikagaku 120491) was then added. Cell suspensions were incubated at 30°C and their optical density (OD600) were recorded at 3 min intervals.
Whole-Genome sequencing
W303-1A and all six evolved clones were streaked for single colonies on 2% YEP Dextrose plates. Single colonies were grown in 2% YEP Dextrose liquid cultures at 30°C and genomic DNA was extracted by spooling as described (Treco, 1987). Paired-end sequencing libraries were created with 5μg of input genomic DNA using the protocol outlined by the Illumina Genomic DNA Sample Prep Kit, except using adapters and reagents purchased individually from various suppliers. Sequencing flow cells were prepared using the Illumina Standard Cluster Generation Kit. Samples were sequenced on the Illumina Genome Analyzer II, and image analysis and data extraction were performed using Illumina RTA 1.5.35.0. Sequence reads were aligned to the S288c reference genome (SGD, as of Feb 2, 2010) using BWA v0.5.7 (Li & Durbin, 2009). Whole-genome pileup files were generated from the aligned sequence data using SAMtools v0.1.7 (Li, et al., 2009) and SNPs were identified using custom filters. Briefly, SNPs passed the filter if they were represented in at least 40% of reads in the evolved strain and at most 10% in the ancestor. Additional heuristic filters included a confirming read from both strands, with at least 5 reads covering the position in both strains, and no more than one ambiguous SNP call (“N”) or deletion (“*”) at that position. All called SNPs were then checked using Sanger sequencing.
Results
Selection of ethanol-tolerant mutants in turbidostat culture
At the conditions used in turbidostat, the maximal growth rate of the parental yeast strain W303-1A in the presence of 8% ethanol was very low and measured as 0.029±0.003 h-1. Selection in the turbidostat was performed in three stages with a gradually increasing concentration in each stage, 6, 7, and finally 8% ethanol (Fig. 1). Each stage resulted in an increase in the apparent growth rate of the culture; at the beginning of stage III (16 days) it was 0.24 h-1, and at the end of experiment (21 days, 141 generations) 0.32 h-1, an increase of an order of magnitude compared to the parental strain.
Fig. 1.
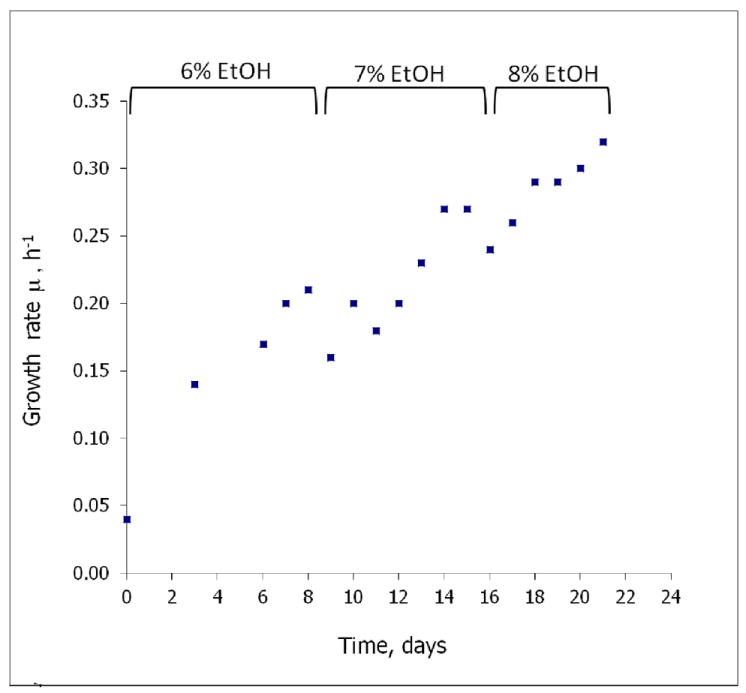
A significant and rapid increase in the growth rate of a yeast population is observed during selection in a turbidostat. The turbidostat was operated in the range of OD600 1.0-2.0 at 30°C in YPD medium containing ethanol 6% (v/v) for stage I, 7% for stage II and 8% for stage III (see text for details). Growth rate (μ) was measured every 24 hours and the dilution rate was adjusted such that D=μ.
A population sample obtained from each of three stages of selection was grown in YPD medium without ethanol in 3 cycles of 24 hours each, ca., 24 generations. When these samples were retested for growth in the presence of ethanol, no significant changes in the apparent growth rate were observed (results not shown), indicating the heritable character of the phenotype.
Evolving continuous cultures may result in clonal interference (Kao & Sherlock, 2008), and we thus assessed the variability of our population. In a random sample of cells from stage III of the turbidostat culture, all cells formed colonies on YPD agar medium supplemented with 7% ethanol. A sample of these cells (105) was randomly tested on solid YPD medium supplemented with 9% ethanol. About 30% of the colonies grew well, whereas ca., 5 % did not grow at all. The other colonies showed intermediate degrees of growth capability (Fig. 2). We chose for further investigation two clones isolated from the stage III population that formed especially dense colonies on 9% ethanol and designated them 9C and 9E (Fig. 2). The maximal growth rates of these clones in liquid medium containing 8% ethanol were superior to that of the W303-1A (μmax = 0.029 ±0.002 h-1), with clone 9C (μmax = 0.117 ±0.003 h-1) growing slightly better than 9E (μmax = 0.102 ±0.002 h-1).
Fig. 2.
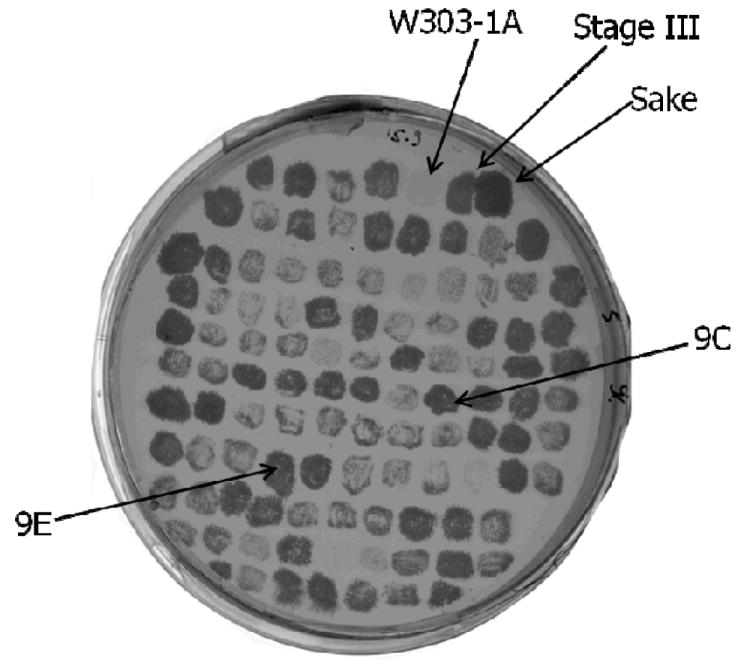
The culture derived from the selection in turbidostat is heterogeneous. A sample of the turbidostat culture from the last stage of selection was streaked and dispersed on solid YPD medium to obtain single colonies. 105 of these colonies were randomly selected and plated onto solid YPD medium supplemented with 9% ethanol and allowed to grow for 48 hours. The arrows point at the following clones: Sake K9, W303-1A, the mixed population of the last selection stage (Stage III), clone 9E and clone 9C that were selected for further investigation.
Frequently, tolerance to a certain stressor involves general stress mechanisms (Piper, 1995, MartinezPastor, et al., 1996) thus resulting in a degree of cross-protection. Indeed, the improved growth rate of 9C and 9E and of the mixed Stage III population in the presence of ethanol was not the only characteristic acquired during the selection in turbidostat. All three were less sensitive to 1 mM H2O2 and to 8 mM caffeine than W303-1A; the mixed population and the clone 9E were more tolerant to caffeine than the clone 9C (Fig.3).
Fig. 3.
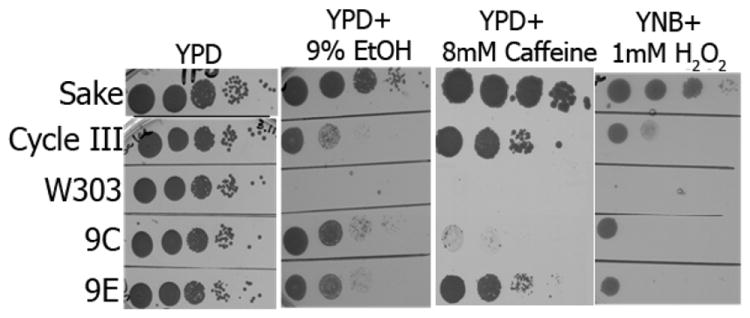
Turbidostat-selected stage III population and clones 9C and 9E are less sensitive to ethanol, caffeine and oxidative stress, compared to parental W303-1A cells. Conditions tested were: 30°C, YPD: 24 h; YPD+9% Ethanol: 72 h; YPD + 8mM Caffeine: 7 days; YNB+ 1mM H2O2: 48h.
A single gene is apparently responsible for ethanol tolerance of clones 9E and 9C
Clones 9E and 9C, as well as the parental W303-1A, were backcrossed to W303-1Aα. The diploid of the parental strain was as sensitive to ethanol as the haploid (not shown). For 9E, the resulting heterozygous diploid was ethanol-tolerant (Fig. 4A, left panel), indicating that ethanol tolerance in this mutant is dominant. In contrast, for 9C, the growth of the resulting heterozygous diploid on ethanol-containing agar was consistent with a semi-dominant phenotype (Fig. 4B, right panel), as it grew better than the parental W303-1A, but not as well as clone 9C itself. Analysis of 6 tetrads from each diploid revealed a 2:2 segregation of the ethanol-tolerant phenotype (Fig. 4), a pattern consistent with the hypothesis that a single allele is responsible for the ethanol-tolerant phenotype.
Fig. 4.
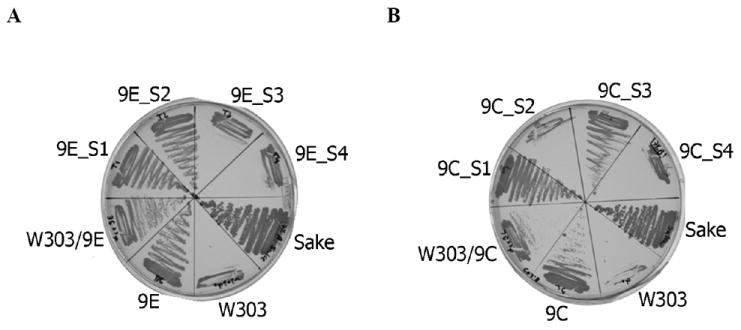
A single gene is responsible for the ability of clones 9C and 9E to proliferate in the presence of high ethanol concentrations. Clones 9E and 9C were mated with W303-1Aα to create diploids W303-1A/9E (Panel A) and W303-1A/9C (Panel B). Following sporulation, the tetrads were dissected. Spores of W303-1A/9E were marked as 9E_S1, 9E_S2, 9E_S3 and 9E_S4. Spores of W303-1A/9C were marked as 9C_S1, 9C_S2, 9C_S3 and 9C_S4. Diploids, as well as resulting pores, were grown on solid YPD+ 7% ethanol. Sake K9 was used as a positive control. The strains were photographed after 4 days in culture at 30°C.
The majority of ethanol-tolerant clones carry mutations in SSD1
We whole-genome sequenced clones 9C and 9E and identified SNPs relative to W303-1A that, presumably, arose during the culture in turbidostat (Table 4). To possibly identify additional mutations of interest, and to further gauge the level of heterogeneity in the turbidostat population, we selected for sequencing four additional ethanol-tolerant clones from earlier turbidostat populations, based on the same criteria that were used for the isolation of clones 9E and 9C (Fig. 2). Clones 1-E2 and 1-G1 were isolated from the sample of stage I population, clones 2-F2 and 2-G2 were isolated from the stage II population.
Table 4.
Mutations detected in the genome of ethanol-tolerant clones derived from the strain W303-1A. Mutations are relative to the Watson strand, rather than the affected feature.
| Clone | Genome modification
|
|||||
|---|---|---|---|---|---|---|
| Chromosome | Position | Gene | Mutation | Amino acid change | ||
| 9C | chrXI | 519838 | UTH1 | G to T | Asp 224 to Tyr | |
| chrXIII | 89994 | RPM2 | C to A | Met 246 to Ile | ||
| chrXI | 622969 | MLP1 | A to T | Ile 1175 to Fhe | ||
| chrXVI | 102249 | ENV7 | C to T | Gly 152to Arg | ||
| chrV | 435785 | Tk(CUU)E2 | A to T | |||
| chrIV | 785204 | TRM82 | G to C | No change | ||
| chrVII | 1014221 | unknown | A to T | |||
|
| ||||||
| 9E | chrXI | 103298 | FAS1 | T to G | Leu 875 to Val | |
| chrIV | 1047297 | SSD1 | T to A | Stop 698 to Leu | ||
| chrIV | 785204 | TRM82 | G to C | No change | ||
|
| ||||||
| 1-E2 | chrIV | 1047297 | SSD1 | T to A | Stop 698 to Leu | |
| chrVII | 448733 | unknown | C to A | |||
| chrXVI | 486441 | unknown | A to T | |||
| chrIV | 785204 | TRM82 | G to C | No change | ||
|
| ||||||
| 1-G1 | chrIV | 1047296 | SSD1 | C to A | Stop 698 to Tyr | |
| chrI | 111917 | CCR4 | G to A | Pro 482 to Ser | ||
| chrXV | 735487 | PTP2 | T to C | Leu 521 to Ser | ||
| chrIV | 785204 | TRM82 | G to C | No change | ||
| chrII | 805593 | MAL32 | C to T | No change | ||
|
| ||||||
| 2-F2 | chrVII | 410268 | RPT6 | A to G | Ile 341 to Thr | |
| chrXIV | 208299 | SIN4 | G to T | Asp 457 to Tyr | ||
| chrIV | 1486864 | SPS1 | T to C | No change | ||
| chrIV | 1047297 | SSD1 | T to A | Stop 698 to Leu | ||
| chrV | 45658 | YEL057C | A to C | Ala 22 to Ser | ||
| chrXVI | 486441 | unknown | A to T | |||
| chrIV | 785204 | TRM82 | G to C | No change | ||
|
| ||||||
| 2-G2 | chrIV | 1047297 | SSD1 | T to A | Stop 698 to Leu | |
| chrXIII | 746464 | DFG5 | G to C | Cys 37 to Ser | ||
| chrXVI | 486441 | unknown | A to T | |||
| chrIV | 785204 | TRM82 | G to C | No change | ||
Mutations identified in the whole genome sequencing of all six clones are shown in Table 4. Although all the clones contained a point mutation in TRM82, we assigned it a low priority for two reasons: (1) it was a silent mutation and (2) clones 9C and 9E showed different segregation patterns (Fig. 4) making it unlikely that the same mutation underlies these patterns. The end justified our premise, since we were able to identify the bona fide mutations (see below) that accounted for ethanol tolerance. Between them, five out of six ethanol-tolerant clones contained in total at least two independent mutations in the SSD1 gene. These mutations result in the replacement of the stop codon at position 698 of SSD1 with an amino acid codon. In four mutants, 1-E2, 2-F2, 2-G2 and 9E, the substitution resulted in the codon for leucine while in the clone 1-G1 the stop codon was substituted for a tyrosine codon.
Involvement of SSD1 in ethanol tolerance
SSD1 is known to be polymorphic within commonly-used S. cerevisiae laboratory strains, with two common genetic variants: the SSD1-V allele that encodes a protein of 1251 amino acids in length, and the ssd1-d allele that terminates the protein at position 698 and encodes a protein of 697 amino acids in length (Sutton, et al., 1991, Jorgensen, et al., 2002).
To determine the effect of the SSD1-V allele (which is identical to SSD1 allele of the clone 1-G1) on ethanol tolerance we cloned SSD1-V from BY4741, in the centromeric plasmid pRS416 (Fig. 5). This construct conferred on W303-1A the ability to grow in the presence of 7% ethanol (BY4741 also has this ability, data not shown).
Fig. 5.
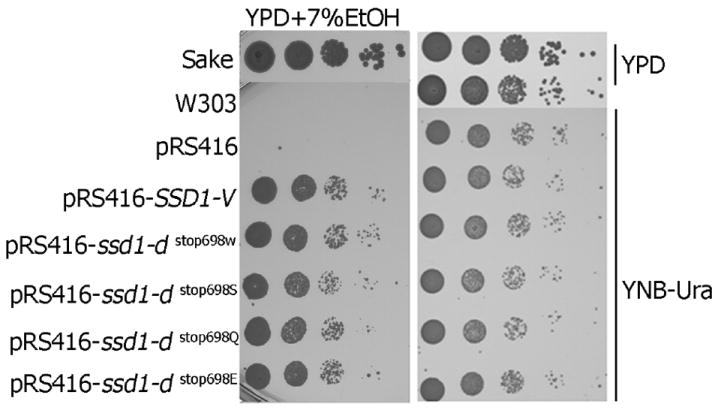
All stop codon substitutions in the ssd1-d allele confer ethanol tolerance similar to the SSD1-V allele. The ssd1-d alleles with the stop codon substituted for one encoding Trp, Ser, Glu or Gln were constructed by site direct mutagenesis and sub-cloned into pRS416. Aliquots (5μl) of 10-fold serial dilutions of Sake, W303-1A and W303-1A transformed with pRS416 (empty plasmid) or pRS416, which carried the indicated ssd1-d variants, were plated onto YPD +7% ethanol and cultured at 30°C for 5 days. Growth of the same strains under unconstrained conditions (without ethanol) was compared, while strains without plasmids were grown on YPD medium and strains with plasmid were grown on YNB-Ura medium.
As part of characterizing the stop codon mutation in SSD1 and of understanding the structure-function relationship in this protein it was important to establish whether a specific amino acid(s) renders the allele functional in ethanol tolerance or just the full length protein is sufficient. Substitution of only one base at the “TAG” stop codon of ssd1-d can results in 5 possible amino acids: leucine, tryptophan, serine, glutamate or glutamine at position 698. We constructed all these variants, except for one with leucine, already found in the 9E mutant, and expressed each of them from the centromeric plasmid pRS416 in W303-1A. All the constructs conferred an ethanol-tolerant phenotype, similar to that of SSD1-V allele (Fig. 5).
We sequenced the SSD1 gene in 13 more clones isolated from the stage III sample that formed dense colonies on 9% ethanol. Ten of these 13 clones contained a mutation of the stop codon in position 698; six contained the codon for lysine, three had leucine and one had tyrosine.
The UTH1 gene is responsible for the ethanol tolerance in clone 9C
Among the six clones subjected to whole genome sequencing (Table 4), clone 9C was the only one that maintained the parental ssd1-d allele of W303-1A. As mentioned above, we mated 9C with W303-1Aα; we sporulated the resulting diploid, and dissected three tetrads. Each tetrad had two ethanol-tolerant spores and two ethanol-sensitive spores (Fig. 4B). We used colony PCR to sequence the genes ENV7, RPM2, MLP1 and UTH1 (Table 4) in the progeny of each spore. We did not genotype mutations in Tk(CUU)E2 and TRM82, which were also segregating in this cross. The UTH1D224Y allele was the only one consistently co-segregated with the ethanol tolerance phenotype.
We deleted uth1Δ in W303-1A, and found that the resulting mutant was more tolerant to ethanol than the parental W303-1A, though more sensitive than the clone 9C (Fig. 6). Thus, the UTH1D224Y allele of the clone 9C was more effective in conferring ethanol tolerance than deletion of UTH1. When the UTH1 WT allele from W303-1A was expressed exogenously in W303-1A uth1Δ, the deletion mutant became as sensitive to ethanol is the wild-type (Fig. 6), suggesting that the deletion mutation is recessive. Thus, elimination of UTH1 confers some ethanol tolerance, but not as well as the mutation UTH1 D224Y. Perhaps, this mutation, while interrupting some functions of the UTH1 product, preserves others that are important for ethanol-tolerant growth. Indeed, when the UTH1 D224Y allele was expressed in W303-1A uth1Δ, the tolerance was maintained and even somewhat improved (Fig. 6). This explains the semi-dominant effect observed in the 9C/W303-1A diploid (Fig. 4). This diploid harbors the defective UTH1 D224Y allele from clone 9C that supports ethanol tolerance, but also the UTH1 WT from W303-1A that causes ethanol sensitivity.
Fig. 6.
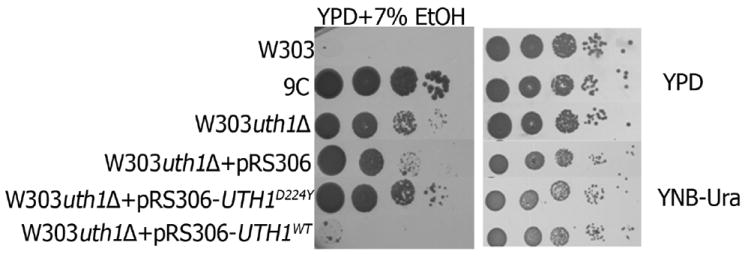
The deletion mutant W303-1A uth1Δ proliferates in the presence of ethanol, but less efficiently than clones expressing the UTH1D224Y mutation. Aliquots (5μl) of 10-fold serial dilutions of W303-1A, W303-1A uth1Δ, W303-1A uth1Δ containing pRS306 vector expressing either UTH1 of W303-1A origin or UTH1D224Y of 9C origin, as well as the clone 9C were plated onto the designated media and cultured at 30°C for 6 days.
In 13 tolerant clones isolated from the sample of stage III, ten contained SSD1 mutations (mentioned above), and two contained UTH1 mutations, one identical to that found in 9C (UTH1D224Y) and one with a different mutation (UTH1N235H). One clone did not contain mutations in either in SSD1 nor in UTH1. Whole genome sequencing of this clone revealed that it contained mutation in three genes: ROT2, SIR4 and DGR2.
Mutations in UTH1 or SSD1 render the W303-1A cell wall more tolerant to zymolyase
The biochemical functions of Ssd1 and Uth1 are unclear, with some reports suggesting that these genes are important for cell wall integrity (Kaeberlein & Guarente, 2002, Reinke, et al., 2004, Ritch, et al., 2010). Deletion of UTH1 confers tolerance against cell wall perturbing agent such as SDS and zymolyase (Ritch, et al., 2010). Sake strains also show strong tolerance to zymolyase as well as to K1 killer toxin, whose target resides in the cell wall (Hara, 1976a, Hara, 1976b). To determine if our ‘tolerant’ clones had enhanced cell wall integrity, we exposed the strains 9C, 9E and W303-1A uth1Δ to zymolyase (Fig. 7).
Fig. 7.
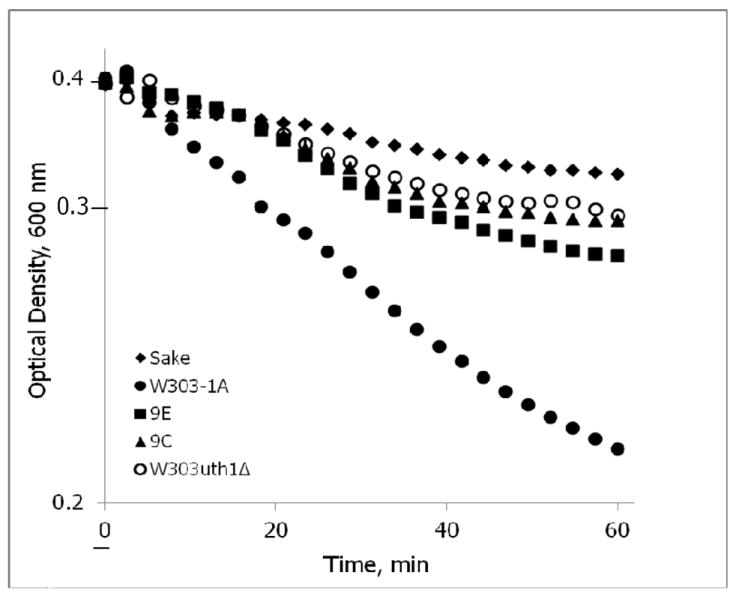
W303-1Auth1Δ, 9C and 9E are more tolerant to zymolyase than the parental W303-1A. Cells of the W303-1Auth1Δ, 9E, 9C, Sake and W303-1A strains, were allowed to grow on YPD media to stationary growth phase, collected by centrifugation, washed three times with deionized water and then resuspended to an OD600 = 0.4 in TE buffer followed by the addition of zymolyase at the time t = 0. The OD600 of cell suspensions was then recorded at 3 min intervals. Assays were performed in triplicate and were highly reproducible; the standard deviation (SD) did not exceed 10% of the mean value.
Treatment with zymolyase in hypotonic buffer causes lysis of yeast, when its cell wall’s mechanical stability is compromised by the digestion of glucan fibers. The time course of digestion is thought to depend upon thickness of the cell wall (Jung, et al., 2005). Following (Ovalle, et al., 1998), we determined the 1st order kinetics of degradation starting with the onset of a stable degradation rate at 15 min and until gradual slowing down 25 min later (Fig. 7). The Sake strain was the most stable among investigated strains with the degradation rate constant of kdeg = 0.166±0.004 h-1 and W303-1A was the least resistant to zymolyase action (kdeg = 0.684±0.007 h-1). The cell wall of 9E (kdeg = 0.420±0.007 h-1), 9C (kdeg = 0.348±0.004 h-1) and of W303-1A uth1Δ (kdeg = 0.302±0.004 h-1) was more robust than that of the W303-1A and less stable than that of the Sake.
Discussion
Our criterion for ethanol tolerance in yeast was the ability of a single cell to form a colony within 6 days on YPD agar containing 7% ethanol. According to this definition, the ethanol-tolerant phenotype appeared in original population of W303-1A, seeded at 106 CFU on the solid YPD+7% ethanol, with a frequency of 5.5±0.5×10-6 (result not shown). Within each stage of selection and between the stages, the average proliferation rate increased (Fig. 1). At none of the stages did the potential for additional selection appear exhausted; the proliferation rate increase did not show any signs of leveling (Fig. 1). In all the clones analyzed so far, a single gene was the major contributor to ethanol tolerance. Perhaps, our decision to collect the population after only 3 weekly cycles of selection prevented us from selecting more complex patterns of adaptation. Nonetheless, these data highlight the potential of this method for practical strain improvement.
Among the best growing clones selected in the turbidostat, the mutation conferring ethanol tolerance that reached the highest frequency was one converting the truncated ssd1-d allele into the full length allele. Polymorphic variants of SSD1 are known to influence cell integrity (Kaeberlein & Guarente, 2002), longevity (Kaeberlein, et al., 2004) and pathogenicity (Wheeler, et al., 2003). W303-1A contains the ssd1-d allele and is sensitive to cell wall perturbing agents such as caffeine and Calcoflour white (Kaeberlein & Guarente, 2002, Reinke, et al., 2004). SSD1 deletion in the S. cerevisiae strain BY4741 renders it ethanol- sensitive (Kubota, et al., 2004, Yoshikawa, et al., 2009).
The second common mutation in our ethanol-tolerant population was in UTH1. UTH1 is a member of the SUN family of fungal genes. Four other fungal SUN genes, SUN4 in S. cerevisiae (Mouassite, et al., 2000), psu1 in S. pombe (Omi, et al., 1999), SUN41 and SIM1/SUN42 in Candida albicans (Firon, et al., 2007) have been implicated in the regulation of the integrity of the yeast cell wall. UTH1 has been identified on the basis of stress tolerance (Bandara, et al., 1998, Camougrand, et al., 2003, Jo, et al., 2008) and longer life span of mutants (Kennedy, et al., 1995). It also interferes with mitochondrial biogenesis (Camougrand, et al., 2000) and is required for the autophagic degradation of mitochondria (Kissova, et al., 2004). Despite these extensive studies, the biochemical function of Uth1p remains unknown. Interestingly, Ritch et al. (Ritch, et al., 2010) demonstrated that deletion of UTH1 (uth1Δ) in the W303 background confers tolerance to cell wall perturbation reagents such as Calcoflour white. Cell wall integrity is frequently considered to be a prerequisite for ethanol tolerance in Sake yeasts (Hara, 1976a, Hara, 1976b).
The UTH1 allele we isolated conferred the ability to grow better on ethanol than either the parental W303-1A or the uth1Δ strain, indicating that the point mutation results in a stronger allele than the deletion with respect to ethanol tolerance. Perhaps in the multiple functions of UTH1 some are beneficial to the ethanol tolerance and some are detrimental. Thus, the point mutation UTH1D224Y cancels the detrimental function, while the complete deletion obliterates both.
There is a connection between the two common mutations conferring ethanol tolerance to W303-1A. Two research groups have shown that the mRNA of UTH1 belongs to a set of Ssd1p-associated mRNAs that are involved in bud morphogenesis and are generally enriched in cell wall related genes (Hogan, et al., 2008, Jansen, et al., 2009). Ssd1 is estimated to have a role in translation of these bound mRNAs. Uth1 is less abundant in cells expressing a functional SSD1 allele (Jansen, et al., 2009).
Although we stressed the difference between the ability of yeast to survive stress and the ability to propagate in the continuous presence of the stress factor, it is obvious that there is an overlap between the two parameters. Thus, cells able to proliferate in the presence of the stressor should, by definition, come up as stress-resistant in some common assays such as growth in the presence of caffeine (‘caffeine stress’) or hydrogen peroxide (‘oxidative stress’). Indeed, the mixed population of the stage III and the clone 9E were considerably more stable than the parental strain in caffeine stress assay (Fig. 3), which is not surprising since caffeine is a known cell wall perturbing agent (Levin, 2005). However, clone 9C was only marginally better than W303-1A. All three samples performed better than W303-1A regarding oxidative stress. Although the link between oxidative stress and the integrity of the cell wall is not obvious, sturdier cell wall may provide better support for yeast outer membrane against oxidative damage.
Cells in which the “general stress response” is induced through stress responsive elements (STREs) activated by the transcription activators Msn2/4, can cope with many stresses (MartinezPastor, et al., 1996). The evolved population, indeed, manifests a degree of cross-protection and therefore the involvement of general stress response pathways cannot be ruled out. Interestingly, none of the clones harbored mutations in the Ras/cAMP/MSN2/4 pathway, known to be responsible for the adaptation to various stresses (MartinezPastor, et al., 1996) and to related changes in growth pattern (Stanhill, et al., 1999). The mechanisms that govern tolerance and are selected in continuous culture may, perhaps, be fundamentally different from mechanisms responsible for stress tolerance in batch culture (possibly due to a different fitness landscape in the turbidostat), though of course it is possible that such mutations are present among the uninvestigated clones at low frequency.
Ethanol exerts multiple effects on the growth of yeast (Zhao & Bai, 2009), ranging from lowering water availability to directly interacting with the cell membrane, DNA and proteins. These effects vary with the stage of fermentation, aeration, temperature, etc. In spite of many efforts (Ma & Liu, 2010) the knowledge obtained so far does not provide direct operative understanding of how ethanol inhibits the growth of yeast. On the other hand, it has to be understood that sequential selection for ethanol tolerance would start with the mutations able to fix a relevant vulnerability of the parental strain. Our results indicate that this vulnerability in W303-1A is a defect in the cell wall. The cell wall probably plays a general role in the stabilization of cell outer membrane either by direct physical support or by interaction with the cell membrane cytoskeleton against the destabilizing effects of ethanol such as membrane expansion (Chen & Engel, 1990). This view is supported by an extraordinary stability of the cell wall of Sake yeast (Fig. 7).
It is clear that the application of the turbidostat as described above has the ability to shed considerable light on the mechanisms of ethanol tolerance, and likely additional relevant mutations will be found in longer term experiments. We also anticipate that it can be used successfully for the improvement of existing industrial strains as it was for laboratory yeast.
Acknowledgments
We wish to thank Mr. A. Grebelsky, Dr. T. Ravid and his group of the Hebrew University for their assistance in providing experimental protocols and overcoming technical difficulties. The funds for this work were provided by the YISSUM R&D Company of the Hebrew University. JWW is supported by the NIH-NIGMS Genetics & Developmental Biology Training Program (NIH GM007790).
References
- Akao T, Yashiro I, Hosoyama A, et al. Whole-Genome Sequencing of Sake Yeast Saccharomyces cerevisiae Kyokai no. 7. DNA Res. 2011;18:423–434. doi: 10.1093/dnares/dsr029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandre H, Ansanay-Galeote V, Dequin S, Blondin B. Global gene expression during short-term ethanol stress in Saccharomyces cerevisiae. Febs Letters. 2001;498:98–103. doi: 10.1016/s0014-5793(01)02503-0. [DOI] [PubMed] [Google Scholar]
- Alper H, Moxley J, Nevoigt E, Fink GR, Stephanopoulos G. Engineering yeast transcription machinery for improved ethanol tolerance and production. Science. 2006;314:1565–1568. doi: 10.1126/science.1131969. [DOI] [PubMed] [Google Scholar]
- Baerends RJS, Qiu JL, Rasmussen S, Nielsen HB, Brandt A. Impaired Uptake and/or Utilization of Leucine by Saccharomyces cerevisiae Is Suppressed by the SPT15-300 Allele of the TATA-Binding Protein Gene. Appl Environ Microb. 2009;75:6055–6061. doi: 10.1128/AEM.00989-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandara PDS, Flattery-O’Brien JA, Grant CM, Dawes IW. Involvement of the Saccharomyces cerevisiae UTH1 gene in the oxidative-stress response. Current Genetics. 1998;34:259–268. doi: 10.1007/s002940050395. [DOI] [PubMed] [Google Scholar]
- Bennett WN, Boraas ME. Isolation of a Fast-Growing Strain of the Rotifer Brachionus-Calyciflorus Pallas Using Turbidostat Culture. Aquaculture. 1988;73:27–36. [Google Scholar]
- Bloom FR, Mcfall E. Isolation and Characterization of D-Serine Deaminase Constitutive Mutants by Utilization of D-Serine as Sole Carbon or Nitrogen-Source. J Bacteriol. 1975;121:1078–1084. doi: 10.1128/jb.121.3.1078-1084.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann CB, Davies A, Cost GJ, Caputo E, Li JC, Hieter P, Boeke JD. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Brown SW, Oliver SG. Isolation of Ethanol-Tolerant Mutants of Yeast by Continuous Selection. Eur J Appl Microbiol Biotechnol. 1982;16:119–122. [Google Scholar]
- Camougrand N, Grelaud-Coq A, Marza E, Priault M, Bessoule JJ, Manon S. The product of the UTH1 gene, required for Bax-induced cell death in yeast, is involved in the response to rapamycin. Mol Microbiol. 2003;47:495–506. doi: 10.1046/j.1365-2958.2003.03311.x. [DOI] [PubMed] [Google Scholar]
- Camougrand NM, Mouassite M, Velours GM, Guerin MG. The “SUN” family: UTH1, an ageing gene, is also involved in the regulation of mitochondria biogenesis in Saccharomyces cerevisiae. Arch Biochem Biophys. 2000;375:154–160. doi: 10.1006/abbi.1999.1655. [DOI] [PubMed] [Google Scholar]
- Chandler M, Stanley GA, Rogers P, Chambers P. A genomic approach to defining the ethanol stress response in the yeast Saccharomyces cerevisiae. Annal Microbiol. 2004;54:427–454. [Google Scholar]
- Chen CH, Engel SG. The Susceptibility of Membrane-Vesicles to Interaction with Ethanol. Chem Phys Lipids. 1990;52:179–187. doi: 10.1016/0009-3084(90)90113-6. [DOI] [PubMed] [Google Scholar]
- Ding JM, Huang XW, Zhang LM, Zhao N, Yang DM, Zhang KQ. Tolerance and stress response to ethanol in the yeast Saccharomyces cerevisiae. Appl Microbiol Biotechnol. 2009;85:253–263. doi: 10.1007/s00253-009-2223-1. [DOI] [PubMed] [Google Scholar]
- Firon A, Aubert S, Iraqui I, et al. The SUN41 and SUN42 genes are essential for cell separation in Candida albicans. Mol Microbiol. 2007;66:1256–1275. doi: 10.1111/j.1365-2958.2007.06011.x. [DOI] [PubMed] [Google Scholar]
- Fujita K, Matsuyama A, Kobayashi Y, Iwahashi H. The genome-wide screening of yeast deletion mutants to identify the genes required for tolerance to ethanol and other alcohols. FEMS Yeast Res. 2006;6:744–750. doi: 10.1111/j.1567-1364.2006.00040.x. [DOI] [PubMed] [Google Scholar]
- Gietz RD, Schiestl RH. Quick and easy yeast transformation using the LiAc/SS carrier DNA/PEG method. Nature Protocols. 2007;2:35–37. doi: 10.1038/nprot.2007.14. [DOI] [PubMed] [Google Scholar]
- Hara S, Sasaki M, Obata T, Nojiro K. Isolation of ethanol-tolerant mutants from sake yeast Kyokai no. 7. J Brew Soc Japan. 1976;71:301–304. [Google Scholar]
- Hara S, Yamamoto N, Fukuda Y, Obata T, Noshiro K. Comparison of physiological characteristics between sake yeast Kyokai no. 7 and its ethanol tolerant mutant. J Brew Soc Japan. 1976;71:564–568. [Google Scholar]
- Harder W, Kuenen JG, Matin A. Microbial Selection in Continuous Culture. J Appl Bacteriol. 1977;43:1–24. doi: 10.1111/j.1365-2672.1977.tb00717.x. [DOI] [PubMed] [Google Scholar]
- Hirasawa T, Yoshikawa K, Nakakura Y, et al. Identification of target genes conferring ethanol stress tolerance to Saccharomyces cerevisiae based on DNA microarray data analysis. J Biotechnol. 2007;131:34–44. doi: 10.1016/j.jbiotec.2007.05.010. [DOI] [PubMed] [Google Scholar]
- Hogan DJ, Riordan DP, Gerber AP, Herschlag D, Brown PO. Diverse RNA-Binding Proteins Interact with Functionally Related Sets of RNAs, Suggesting an Extensive Regulatory System. Plos Biol. 2008;6:2297–2313. doi: 10.1371/journal.pbio.0060255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen JM, Wanless AG, Seidel CW, Weiss EL. Cbk1 regulation of the RNA-binding protein Ssd1 integrates cell fate with translational control. Curr Biol. 2009;19:2114–2120. doi: 10.1016/j.cub.2009.10.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez J, Benitez T. Selection of Ethanol-Tolerant Yeast Hybrids in Ph-Regulated Continuous Culture. Appl Environ Microbiol. 1988;54:917–922. doi: 10.1128/aem.54.4.917-922.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez J, Benitez T. Selection of Ethanol-Tolerant Yeast Hybrids in pH-Regulated Continuous Culture. Appl Environ Microbiol. 1988;54:917–922. doi: 10.1128/aem.54.4.917-922.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo WJ, Loguinov A, Chang M, et al. Identification of genes involved in the toxic response of Saccharomyces cerevisiae against iron and copper overload by parallel analysis of deletion mutants (vol 101, pg 140, 2008) Toxicol Sci. 2008;102:205–205. doi: 10.1093/toxsci/kfm226. [DOI] [PubMed] [Google Scholar]
- Jorgensen P, Nelson B, Robinson MD, Chen YQ, Andrews B, Tyers M, Boone C. High-resolution genetic mapping with ordered arrays of Saccharomyces cerevisiae deletion mutants. Genetics. 2002;162:1091–1099. doi: 10.1093/genetics/162.3.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung WH, Wam P, Ragni E, Popolo L, Nunn CD, Turner MP, Stateva L. Deletion of PDE2, the gene encoding the high-affinity cAMP phosphodiesterase, results in changes of the cell wall and membrane in Candida albicans. Yeast. 2005;22:285–294. doi: 10.1002/yea.1199. [DOI] [PubMed] [Google Scholar]
- Kaeberlein M, Guarente L. Saccharoyrayces cerevisiae MPT5 and SSD1 function in parallel pathways to promote cell wall integrity. Genetics. 2002;160:83–95. doi: 10.1093/genetics/160.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M, Andalis AA, Liszt GB, Fink GR, Guarente L. Saccharomyces cerevisiae SSD1-V confers longevity by a Sir2p-independent mechanism. Genetics. 2004;166:1661–1672. doi: 10.1534/genetics.166.4.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao KC, Sherlock G. Molecular characterization of clonal interference during adaptive evolution in asexual populations of Saccharomyces cerevisiae. Nature Genetics. 2008;40:1499–1504. doi: 10.1038/ng.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy BK, Austriaco NR, Zhang JS, Guarente L. Mutation in the Silencing Gene Sir4 Can Delay Aging in Saccharomyces-Cerevisiae. Cell. 1995;80:485–496. doi: 10.1016/0092-8674(95)90499-9. [DOI] [PubMed] [Google Scholar]
- Kim J, Alizadeh P, Harding T, HefnerGravink A, Klionsky DJ. Disruption of the yeast ATH1 gene confers better survival after dehydration, freezing, and ethanol shock: Potential commercial applications. Appl Environ Microbiol. 1996;62:1563–1569. doi: 10.1128/aem.62.5.1563-1569.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kissova I, Deffieu M, Manon S, Camougrand N. Uth1p is involved in the autophagic degradation of mitochondria. J Biol Chem. 2004;279:39068–39074. doi: 10.1074/jbc.M406960200. [DOI] [PubMed] [Google Scholar]
- Kopp M, Hermisson J. The genetic basis of phenotypic adaptation II: the distribution of adaptive substitutions in the moving optimum model. Genetics. 2009;183:1453–1476. doi: 10.1534/genetics.109.106195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota S, Takeo I, Kume K, et al. Effect of ethanol on cell growth of budding yeast: Genes that are important for cell growth in the presence of ethanol. Biosci Biotechnol Biochem. 2004;68:968–972. doi: 10.1271/bbb.68.968. [DOI] [PubMed] [Google Scholar]
- Lang GI, Murray AW. Estimating the per-base-pair mutation rate in the yeast Saccharomyces cerevisiae. Genetics. 2008;178:67–82. doi: 10.1534/genetics.107.071506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin DE. Cell wall integrity signaling in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 2005;69:262–291. doi: 10.1128/MMBR.69.2.262-291.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Tanaka S. Ethanol fermentation from biomass resources: current state and prospects. Appl Microbiol Biotechnol. 2006;69:627–642. doi: 10.1007/s00253-005-0229-x. [DOI] [PubMed] [Google Scholar]
- Ma MG, Liu ZL. Mechanisms of ethanol tolerance in Saccharomyces cerevisiae. Appl Microbiol Biotechnol. 2010;87:829–845. doi: 10.1007/s00253-010-2594-3. [DOI] [PubMed] [Google Scholar]
- MartinezPastor MT, Marchler G, Schuller C, MarchlerBauer A, Ruis H, Estruch F. The Saccharomyces cerevisiae zinc finger proteins Msn2p and Msn4p are required for transcriptional induction through the stress-response element (STRE) EMBO J. 1996;15:2227–2235. [PMC free article] [PubMed] [Google Scholar]
- Mouassite M, Camougrand N, Schwob E, Demaison G, Laclau M, Guerin M. The ‘SUN’ family: yeast SUN4/SCW3 is involved in cell septation. Yeast. 2000;16:905–919. doi: 10.1002/1097-0061(200007)16:10<905::AID-YEA584>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- Nakazawa N, Harashima S, Oshima Y. Genetic mechanism of incompetence of sporulation of Japanese sake yeast, Kyokai no. 7. J Brew Sci. 1993;88:354–357. [Google Scholar]
- Ogawa Y, Nitta A, Uchiyama H, Imamura T, Shimoi H, Ito K. Tolerance mechanism of the ethanol-tolerant mutant of sake yeast. J Biosci Bioeng. 2000;90:313–320. doi: 10.1016/s1389-1723(00)80087-0. [DOI] [PubMed] [Google Scholar]
- Omi K, Sonoda H, Nagata K, Sugita K. Cloning and characterization of psu1(+), a new essential fission yeast gene involved in cell wall synthesis. Biochem Biophys Res Commun. 1999;262:368–374. doi: 10.1006/bbrc.1999.1209. [DOI] [PubMed] [Google Scholar]
- Ovalle R, Lim ST, Holder B, Jue CK, Moore CW, Lipke PN. A spheroplast rate assay for determination of cell wall integrity in yeast. Yeast. 1998;14:1159–1166. doi: 10.1002/(SICI)1097-0061(19980930)14:13<1159::AID-YEA317>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Piper PW. The heat shock and ethanol stress responses of yeast exhibit extensive similarity and functional overlap. FEMS Microbiol Lett. 1995;134:121–127. doi: 10.1111/j.1574-6968.1995.tb07925.x. [DOI] [PubMed] [Google Scholar]
- Reinke A, Anderson S, McCaffery JM, et al. TOR complex 1 includes a novel component, Tco89p (YPL180w), and cooperates with Ssd1p to maintain cellular integrity in Saccharomyces cerevisiae. J Biol Chem. 2004;279:14752–14762. doi: 10.1074/jbc.M313062200. [DOI] [PubMed] [Google Scholar]
- Ritch JJ, Davidson SM, Sheehan JJ, Austriaco OPN. The Saccharomyces SUN gene, UTH1, is involved in cell wall biogenesis. FEMS Yeast Res. 2010;10:168–176. doi: 10.1111/j.1567-1364.2009.00601.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez OJ, Cardona CA. Trends in biotechnological production of fuel ethanol from different feedstocks. Biores Technol. 2008;99:5270–5295. doi: 10.1016/j.biortech.2007.11.013. [DOI] [PubMed] [Google Scholar]
- Sharma SC. A possible role of trehalose in osmotolerance and ethanol tolerance in Saccharomyces cerevisiae. FEMS Microbiol Lett. 1997;152:11–15. doi: 10.1111/j.1574-6968.1997.tb10402.x. [DOI] [PubMed] [Google Scholar]
- Sherman F, Hicks J. Micromanipulation and Dissection of Asci. Meth Enzymol. 1991;194:21–37. doi: 10.1016/0076-6879(91)94005-w. [DOI] [PubMed] [Google Scholar]
- Shobayashi M, Ukena E, Fujii T, Iefuji H. Genome-wide expression profile of sake brewing yeast under shaking and static conditions. Biosci Biotechnol Biochem. 2007;71:323–335. doi: 10.1271/bbb.60190. [DOI] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A System of Shuttle Vectors and Yeast Host Strains Designed for Efficient Manipulation of DNA in Saccharomyces-Cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanhill A, Schick N, Engelberg D. The yeast Ras/cyclic AMP pathway induces invasive growth by suppressing the cellular stress response. Mol Cell Biol. 1999;19:7529–7538. doi: 10.1128/mcb.19.11.7529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley D, Fraser S, Chambers PJ, Rogers P, Stanley GA. Generation and characterisation of stable ethanol-tolerant mutants of Saccharomyces cerevisiae. J Ind Microbiol Biotechnol. 2010;37:139–149. doi: 10.1007/s10295-009-0655-3. [DOI] [PubMed] [Google Scholar]
- Sutton A, Immanuel D, Arndt KT. The Sit4 Protein Phosphatase Functions in Late G1 for Progression into S-Phase. Mol Cell Biol. 1991;11:2133–2148. doi: 10.1128/mcb.11.4.2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi T, Shimoi H, Ito K. Identification of genes required for growth under ethanol stress using transposon mutagenesis in Saccharomyces cerevisiae. Mol Genet Genom. 2001;265:1112–1119. doi: 10.1007/s004380100510. [DOI] [PubMed] [Google Scholar]
- Treco DA. Preparation of yeast DNA. In: Ausubel FM, B R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K, editors. Current protocols in molecular biology. Greene Publishing and Wiley-Interscience; New York: 1987. pp. 13.11.11–13.11.14. [Google Scholar]
- van Voorst F, Houghton-Larsen J, Jonson L, Kielland-Brandt MC, Brandt A. Genome-wide identification of genes required for growth of Saccharomyces cerevisiae under ethanol stress. Yeast. 2006;23:351–359. doi: 10.1002/yea.1359. [DOI] [PubMed] [Google Scholar]
- Watson TG. Present Status and Future Prospects of Turbidostat. J Appl Chem Biotechnol. 1972;22:229. [Google Scholar]
- Wheeler RT, Kupiec M, Magnelli P, Abeijon C, Fink GR. A Saccharomyces cerevisiae mutant with increased virulence. Proce Natl Acad Sci USA. 2003;100:2766–2770. doi: 10.1073/pnas.0437995100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaji K, Hara S, Mizoguchi H. Influence of ras function on ethanol stress response of sake yeast. J Biosci Bioeng. 2003;96:474–480. doi: 10.1016/S1389-1723(03)70134-0. [DOI] [PubMed] [Google Scholar]
- Yazawa H, Iwahashi H, Uemura H. Disruption of URA7 and GAL6 improves the ethanol tolerance and fermentation capacity of Saccharomyces cerevisiae. Yeast. 2007;24:551–560. doi: 10.1002/yea.1492. [DOI] [PubMed] [Google Scholar]
- Yoshikawa K, Tanaka T, Furusawa C, Nagahisa K, Hirasawa T, Shimizu H. Comprehensive phenotypic analysis for identification of genes affecting growth under ethanol stress in Saccharomyces cerevisiae. FEMS Yeast Res. 2009;9:32–44. doi: 10.1111/j.1567-1364.2008.00456.x. [DOI] [PubMed] [Google Scholar]
- Zhao XQ, Bai FW. Mechanisms of yeast stress tolerance and its manipulation for efficient fuel ethanol production. J Biotechnol. 2009;144:23–30. doi: 10.1016/j.jbiotec.2009.05.001. [DOI] [PubMed] [Google Scholar]


