Abstract
The title compound, C34H54N2O2·0.25H2O, the organic molecule, a potential tetradentate ligand with a bulky phenolic donor, has overall mirror symmetry. A partially occupied water molecule of solvation is present in the lattice. The six-membered 1,3-diazinane ring displays a chair conformation. An intramolecular O—H⋯N hydrogen bond ocurs. In the crystal, molecules are linked by O—H⋯O interactions.
Related literature
For aminobisphenolato ligands in coordination chemistry, see: Wichmann et al. (2012 ▶). For applications of their metal complexes, see: Barroso et al. (2010 ▶); Wong et al. (2010 ▶); Kannan et al. (2008 ▶); Pang et al. (2008 ▶); Tshuva et al. (2001 ▶). For background to the synthetic procedure and related structures, see: Hancock et al. (2011 ▶); Manna et al. (2008 ▶); Mohanty et al. (2008 ▶); Guo et al. (2003 ▶).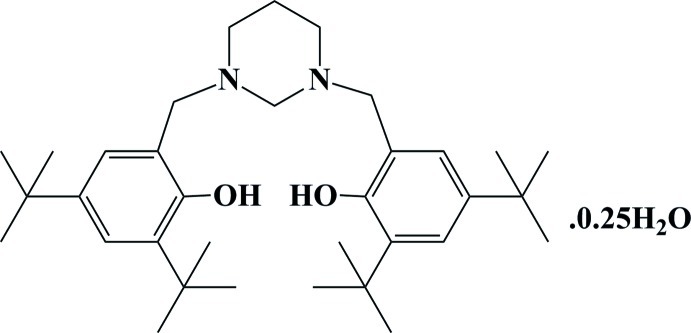
Experimental
Crystal data
C34H54N2O2·0.25H2O
M r = 527.30
Orthorhombic,

a = 8.7292 (8) Å
b = 37.428 (3) Å
c = 10.1806 (9) Å
V = 3326.1 (5) Å3
Z = 4
Mo Kα radiation
μ = 0.07 mm−1
T = 296 K
0.33 × 0.24 × 0.16 mm
Data collection
Bruker SMART APEXII CCD diffractometer
29196 measured reflections
3908 independent reflections
3411 reflections with I > 2σ(I)
R int = 0.031
Refinement
R[F 2 > 2σ(F 2)] = 0.046
wR(F 2) = 0.130
S = 1.05
3908 reflections
180 parameters
1 restraint
H-atom parameters constrained
Δρmax = 0.38 e Å−3
Δρmin = −0.31 e Å−3
Data collection: APEX2 (Bruker, 2009 ▶); cell refinement: SAINT (Bruker, 2009 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: SHELXTL (Sheldrick, 2008 ▶); software used to prepare material for publication: publCIF (Westrip, 2010 ▶) and PLATON (Spek, 2009 ▶).
Supplementary Material
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S1600536812026505/fj2558sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536812026505/fj2558Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O1—H1⋯N1 | 0.82 | 2.00 | 2.6880 (13) | 142 |
| O2—H2⋯O1i | 0.85 | 2.22 | 3.036 (5) | 161 |
Symmetry code: (i)  .
.
Acknowledgments
Financial support from the Jiangsu Key Laboratory for Environment Functional Materials, a project funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD) and the Innovation Program for graduate students of USTS is gratefully acknowledged.
supplementary crystallographic information
Comment
In coordination chemistry, various ligands are used to control the environment of the metal. Especially, the electronic and steric properties of ligands are used to control the reactivity of metal species. Aminobisphenolato ligand is attractive, because its substituents at the phenolate rings as well as the position and nature of the side chain donor are easily tuneable features; thus different electronic and steric properties are available (Wichmann et al. 2012). Such metal complexes have been found to display catalytic activity for polymerization of cyclic esters (Hancock et al., 2011) (biodegradable polymers) and polymerization of olefins (Tshuva et al., 2001). They can also catalyse Tischenko reactions (Pang et al., 2008), sulfoxidations (Barroso et al., 2010), olefin epoxidation (Wong et al., 2010), hydrogenation of ketones (Kannan et al., 2008) and Mizorokie-Heck coupling reaction (Mohanty et al., 2008). Generally, aminobisphenolato ligands are prepared by Mannich condensation from formaldehyde, phenol and a primary amine (Manna et al., 2008; Guo et al., 2003). Herein we present a new ligand, in which two substituted phenols are bridge-linked by a tetrahydropyrimidine ring. The molecules form a mirror symmetric structure, as illustrated in Scheme 1.
In the title compound, the C—N bond distances are between 1.4580 (14) to 1.4763 (15) Å whereas the bond length of C—O is 1.3763 (15) Å. The bond angles around the nitrogen atoms range from 110.35 (12)o to 112.08 (10)o, which is in agreement with those in similar structure (Guo et al., 2003). Two phenolate groups are linked by a tetrahydropyrimidine ring. The overall geometry is mirror-symmetric (Fig.1). The six-member 1,3-diazacyclohexane ring displays in a chair-configuration. Compound molecules were stabilized by hydrogen bonds including intra-molecular O—H···N interaction and inter-molecular O—H···O interaction (Fig. 2).
Experimental
The title compound was prepared as follows. To a solution of 2,4-di-butylphenol (24.77 g, 0.12 mol) in 20 ml of methanol was added 7 ml of formaldehyde(0.08 mol) and 2.0 ml 1,3-propanediamine. The mixture was refluxed for 3 d at 65 oC. In the process white precipitates were produced gradually. After being filtered and washed with methanol for 3 times, white product of C136H218N8O9 was obtained in a yield of 90.7% (based on diamine). Single crystals were grown from ethyl acetate, m.p. = 185 °C, Anal. calcd for C136H218N8O9: C, 77.44; H, 10.42; N, 5.31; Found: C, 77.02; H, 10.55; N, 5.27. IR(KBr, cm-1) 3434(w), 2956(s), 2906(s), 2870(s), 2806(s), 2726(m), 2680(m), 1607(m), 1480(s), 1459(s), 1442(s), 1392(m), 1362(s), 1307(s), 1286(w), 1235(s), 1204(m), 1189(m), 1167(m), 1123(w), 1110(m), 1095(m), 989(m), 883(m), 822(w), 797(w), 761(w), 724(w), 682(w), 460(w).
Refinement
Tertiary Carbon H atoms were constrained to ideal geometry, with C—H = 0.98 Å and Uiso(H) = 1.5Ueq(C), All other H atoms were placed in geometrically idealized positions and constrained to ride on their parent atoms, with C—H = 0.93 (aromatic and alkenyl) Uiso(H) = 1.2Ueq(C). The dispalcement parameters for the water O atom were very large at full occupancy. When refined, its fractional occupancy converged to close to 0.25 and was then set at this value.
Figures
Fig. 1.
The molecular structure of the title compound with ellipsoids scaled to 30% probability.
Fig. 2.
intra and inter molecular contacts (dashed line) as well as molecular packing of the title compound along c axis.
Crystal data
| C34H54N2O2·0.25H2O | F(000) = 1162 |
| Mr = 527.30 | Dx = 1.053 Mg m−3 |
| Orthorhombic, Pnma | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2ac 2n | Cell parameters from 9920 reflections |
| a = 8.7292 (8) Å | θ = 2.6–27.6° |
| b = 37.428 (3) Å | µ = 0.07 mm−1 |
| c = 10.1806 (9) Å | T = 296 K |
| V = 3326.1 (5) Å3 | Prism, colorless |
| Z = 4 | 0.33 × 0.24 × 0.16 mm |
Data collection
| Bruker SMART APEXII CCD diffractometer | 3411 reflections with I > 2σ(I) |
| Radiation source: fine-focus sealed tube | Rint = 0.031 |
| Graphite monochromator | θmax = 27.6°, θmin = 1.6° |
| phi and ω scans | h = −11→11 |
| 29196 measured reflections | k = −48→48 |
| 3908 independent reflections | l = −13→13 |
Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.046 | H-atom parameters constrained |
| wR(F2) = 0.130 | w = 1/[σ2(Fo2) + (0.0634P)2 + 1.2767P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.05 | (Δ/σ)max = 0.038 |
| 3908 reflections | Δρmax = 0.38 e Å−3 |
| 180 parameters | Δρmin = −0.31 e Å−3 |
| 1 restraint | Extinction correction: SHELXL97 (Sheldrick, 2008), Fc*=kFc[1+0.001xFc2λ3/sin(2θ)]-1/4 |
| Primary atom site location: structure-invariant direct methods | Extinction coefficient: 0.0034 (7) |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | Occ. (<1) | |
| N1 | 0.24037 (12) | 0.78185 (2) | 0.40084 (10) | 0.0226 (2) | |
| O1 | 0.50878 (10) | 0.81077 (2) | 0.47464 (9) | 0.0290 (2) | |
| H1 | 0.4572 | 0.7956 | 0.4369 | 0.044* | |
| O2 | 0.7322 (8) | 0.7500 | 0.5233 (7) | 0.0500 (15)* | 0.25 |
| H2 | 0.6880 | 0.7299 | 0.5133 | 0.075* | 0.25 |
| C1 | 0.44073 (13) | 0.84373 (3) | 0.46035 (11) | 0.0218 (2) | |
| C2 | 0.30352 (13) | 0.84657 (3) | 0.38874 (11) | 0.0224 (2) | |
| C3 | 0.23273 (13) | 0.87966 (3) | 0.37546 (12) | 0.0228 (2) | |
| H3A | 0.1408 | 0.8812 | 0.3297 | 0.027* | |
| C4 | 0.29645 (13) | 0.91060 (3) | 0.42917 (11) | 0.0215 (2) | |
| C5 | 0.43413 (13) | 0.90681 (3) | 0.49783 (11) | 0.0220 (2) | |
| H5A | 0.4783 | 0.9272 | 0.5335 | 0.026* | |
| C6 | 0.50974 (13) | 0.87414 (3) | 0.51622 (11) | 0.0211 (2) | |
| C7 | 0.21487 (13) | 0.94668 (3) | 0.41266 (12) | 0.0248 (3) | |
| C8 | 0.05775 (16) | 0.94463 (4) | 0.47962 (16) | 0.0397 (3) | |
| H8A | 0.0056 | 0.9671 | 0.4697 | 0.060* | |
| H8B | 0.0712 | 0.9395 | 0.5713 | 0.060* | |
| H8C | −0.0019 | 0.9260 | 0.4397 | 0.060* | |
| C9 | 0.19124 (19) | 0.95464 (4) | 0.26617 (16) | 0.0429 (4) | |
| H9A | 0.2890 | 0.9560 | 0.2232 | 0.064* | |
| H9B | 0.1386 | 0.9770 | 0.2565 | 0.064* | |
| H9C | 0.1314 | 0.9359 | 0.2272 | 0.064* | |
| C10 | 0.30511 (17) | 0.97759 (4) | 0.4729 (2) | 0.0484 (4) | |
| H10A | 0.4038 | 0.9792 | 0.4317 | 0.073* | |
| H10B | 0.3180 | 0.9735 | 0.5654 | 0.073* | |
| H10C | 0.2503 | 0.9995 | 0.4596 | 0.073* | |
| C11 | 0.66220 (13) | 0.87198 (3) | 0.59218 (11) | 0.0240 (3) | |
| C12 | 0.71429 (15) | 0.90887 (4) | 0.64144 (14) | 0.0334 (3) | |
| H12A | 0.6375 | 0.9187 | 0.6985 | 0.050* | |
| H12B | 0.7290 | 0.9245 | 0.5678 | 0.050* | |
| H12C | 0.8089 | 0.9065 | 0.6887 | 0.050* | |
| C13 | 0.64494 (16) | 0.84776 (4) | 0.71373 (13) | 0.0365 (3) | |
| H13A | 0.5661 | 0.8571 | 0.7698 | 0.055* | |
| H13B | 0.7401 | 0.8470 | 0.7609 | 0.055* | |
| H13C | 0.6178 | 0.8241 | 0.6862 | 0.055* | |
| C14 | 0.78871 (14) | 0.85726 (4) | 0.50186 (12) | 0.0299 (3) | |
| H14A | 0.7992 | 0.8725 | 0.4265 | 0.045* | |
| H14B | 0.7619 | 0.8336 | 0.4738 | 0.045* | |
| H14C | 0.8840 | 0.8565 | 0.5489 | 0.045* | |
| C15 | 0.23842 (15) | 0.81429 (3) | 0.31813 (12) | 0.0261 (3) | |
| H15A | 0.1338 | 0.8193 | 0.2919 | 0.031* | |
| H15B | 0.2976 | 0.8099 | 0.2391 | 0.031* | |
| C16 | 0.2173 (2) | 0.7500 | 0.32086 (16) | 0.0228 (3) | |
| H16A | 0.2891 | 0.7500 | 0.2481 | 0.027* | |
| H16B | 0.1143 | 0.7500 | 0.2851 | 0.027* | |
| C17 | 0.12559 (16) | 0.78340 (3) | 0.50660 (13) | 0.0310 (3) | |
| H17A | 0.0237 | 0.7850 | 0.4690 | 0.037* | |
| H17B | 0.1426 | 0.8045 | 0.5601 | 0.037* | |
| C18 | 0.1380 (3) | 0.7500 | 0.59142 (19) | 0.0369 (4) | |
| H18A | 0.2354 | 0.7500 | 0.6373 | 0.044* | |
| H18B | 0.0569 | 0.7500 | 0.6565 | 0.044* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| N1 | 0.0289 (5) | 0.0148 (4) | 0.0242 (5) | −0.0005 (3) | −0.0016 (4) | −0.0007 (3) |
| O1 | 0.0287 (4) | 0.0196 (4) | 0.0387 (5) | 0.0034 (3) | −0.0068 (4) | 0.0017 (3) |
| C1 | 0.0231 (5) | 0.0188 (5) | 0.0234 (5) | 0.0019 (4) | 0.0001 (4) | 0.0021 (4) |
| C2 | 0.0261 (6) | 0.0176 (5) | 0.0235 (5) | −0.0020 (4) | −0.0028 (4) | 0.0005 (4) |
| C3 | 0.0220 (5) | 0.0199 (5) | 0.0265 (6) | −0.0006 (4) | −0.0044 (4) | 0.0013 (4) |
| C4 | 0.0203 (5) | 0.0189 (5) | 0.0252 (6) | −0.0002 (4) | 0.0010 (4) | −0.0002 (4) |
| C5 | 0.0211 (5) | 0.0205 (5) | 0.0242 (5) | −0.0030 (4) | 0.0009 (4) | −0.0028 (4) |
| C6 | 0.0195 (5) | 0.0244 (5) | 0.0195 (5) | −0.0009 (4) | 0.0000 (4) | 0.0006 (4) |
| C7 | 0.0225 (5) | 0.0167 (5) | 0.0352 (6) | 0.0008 (4) | −0.0001 (5) | −0.0008 (4) |
| C8 | 0.0311 (7) | 0.0340 (7) | 0.0540 (9) | 0.0073 (5) | 0.0090 (6) | 0.0041 (6) |
| C9 | 0.0556 (9) | 0.0304 (6) | 0.0429 (8) | 0.0106 (6) | 0.0013 (7) | 0.0108 (6) |
| C10 | 0.0369 (8) | 0.0217 (6) | 0.0866 (13) | 0.0031 (5) | −0.0134 (8) | −0.0140 (7) |
| C11 | 0.0202 (5) | 0.0308 (6) | 0.0210 (5) | 0.0006 (4) | −0.0017 (4) | 0.0005 (4) |
| C12 | 0.0252 (6) | 0.0406 (7) | 0.0345 (7) | −0.0015 (5) | −0.0065 (5) | −0.0092 (6) |
| C13 | 0.0314 (7) | 0.0525 (8) | 0.0257 (6) | −0.0013 (6) | −0.0032 (5) | 0.0105 (6) |
| C14 | 0.0218 (6) | 0.0394 (7) | 0.0285 (6) | 0.0023 (5) | −0.0008 (5) | −0.0026 (5) |
| C15 | 0.0343 (6) | 0.0166 (5) | 0.0274 (6) | −0.0020 (4) | −0.0081 (5) | 0.0016 (4) |
| C16 | 0.0291 (8) | 0.0159 (7) | 0.0233 (8) | 0.000 | −0.0030 (6) | 0.000 |
| C17 | 0.0370 (7) | 0.0227 (6) | 0.0333 (7) | 0.0019 (5) | 0.0059 (5) | −0.0046 (5) |
| C18 | 0.0523 (12) | 0.0298 (9) | 0.0286 (9) | 0.000 | 0.0117 (9) | 0.000 |
Geometric parameters (Å, º)
| N1—C16 | 1.4575 (13) | C10—H10A | 0.9600 |
| N1—C17 | 1.4719 (16) | C10—H10B | 0.9600 |
| N1—C15 | 1.4777 (14) | C10—H10C | 0.9600 |
| O1—C1 | 1.3770 (13) | C11—C14 | 1.5390 (17) |
| O1—H1 | 0.8200 | C11—C12 | 1.5379 (17) |
| O2—H2 | 0.8500 | C11—C13 | 1.5414 (17) |
| C1—C2 | 1.4062 (16) | C12—H12A | 0.9600 |
| C1—C6 | 1.4078 (15) | C12—H12B | 0.9600 |
| C2—C3 | 1.3909 (15) | C12—H12C | 0.9600 |
| C2—C15 | 1.5162 (15) | C13—H13A | 0.9600 |
| C3—C4 | 1.3962 (15) | C13—H13B | 0.9600 |
| C3—H3A | 0.9300 | C13—H13C | 0.9600 |
| C4—C5 | 1.3975 (16) | C14—H14A | 0.9600 |
| C4—C7 | 1.5359 (15) | C14—H14B | 0.9600 |
| C5—C6 | 1.4023 (16) | C14—H14C | 0.9600 |
| C5—H5A | 0.9300 | C15—H15A | 0.9700 |
| C6—C11 | 1.5414 (15) | C15—H15B | 0.9700 |
| C7—C10 | 1.5283 (17) | C16—N1i | 1.4575 (13) |
| C7—C9 | 1.535 (2) | C16—H16A | 0.9700 |
| C7—C8 | 1.5335 (18) | C16—H16B | 0.9700 |
| C8—H8A | 0.9600 | C17—C18 | 1.5231 (16) |
| C8—H8B | 0.9600 | C17—H17A | 0.9700 |
| C8—H8C | 0.9600 | C17—H17B | 0.9700 |
| C9—H9A | 0.9600 | C18—C17i | 1.5231 (16) |
| C9—H9B | 0.9600 | C18—H18A | 0.9700 |
| C9—H9C | 0.9600 | C18—H18B | 0.9700 |
| C16—N1—C17 | 110.29 (10) | C14—C11—C13 | 109.83 (10) |
| C16—N1—C15 | 110.62 (9) | C12—C11—C13 | 107.17 (10) |
| C17—N1—C15 | 112.13 (9) | C14—C11—C6 | 109.79 (9) |
| C1—O1—H1 | 109.5 | C12—C11—C6 | 111.83 (10) |
| O1—C1—C2 | 119.33 (10) | C13—C11—C6 | 110.44 (10) |
| O1—C1—C6 | 119.81 (10) | C11—C12—H12A | 109.5 |
| C2—C1—C6 | 120.85 (10) | C11—C12—H12B | 109.5 |
| C3—C2—C1 | 119.74 (10) | H12A—C12—H12B | 109.5 |
| C3—C2—C15 | 119.80 (10) | C11—C12—H12C | 109.5 |
| C1—C2—C15 | 120.32 (10) | H12A—C12—H12C | 109.5 |
| C2—C3—C4 | 121.57 (10) | H12B—C12—H12C | 109.5 |
| C2—C3—H3A | 119.2 | C11—C13—H13A | 109.5 |
| C4—C3—H3A | 119.2 | C11—C13—H13B | 109.5 |
| C3—C4—C5 | 117.02 (10) | H13A—C13—H13B | 109.5 |
| C3—C4—C7 | 120.11 (10) | C11—C13—H13C | 109.5 |
| C5—C4—C7 | 122.87 (10) | H13A—C13—H13C | 109.5 |
| C4—C5—C6 | 124.06 (10) | H13B—C13—H13C | 109.5 |
| C4—C5—H5A | 118.0 | C11—C14—H14A | 109.5 |
| C6—C5—H5A | 118.0 | C11—C14—H14B | 109.5 |
| C5—C6—C1 | 116.73 (10) | H14A—C14—H14B | 109.5 |
| C5—C6—C11 | 121.28 (10) | C11—C14—H14C | 109.5 |
| C1—C6—C11 | 121.99 (10) | H14A—C14—H14C | 109.5 |
| C10—C7—C9 | 108.21 (12) | H14B—C14—H14C | 109.5 |
| C10—C7—C8 | 108.69 (11) | N1—C15—C2 | 112.34 (9) |
| C9—C7—C8 | 108.76 (11) | N1—C15—H15A | 109.1 |
| C10—C7—C4 | 112.50 (10) | C2—C15—H15A | 109.1 |
| C9—C7—C4 | 109.83 (10) | N1—C15—H15B | 109.1 |
| C8—C7—C4 | 108.78 (10) | C2—C15—H15B | 109.1 |
| C7—C8—H8A | 109.5 | H15A—C15—H15B | 107.9 |
| C7—C8—H8B | 109.5 | N1—C16—N1i | 109.75 (13) |
| H8A—C8—H8B | 109.5 | N1—C16—H16A | 109.7 |
| C7—C8—H8C | 109.5 | N1i—C16—H16A | 109.7 |
| H8A—C8—H8C | 109.5 | N1—C16—H16B | 109.7 |
| H8B—C8—H8C | 109.5 | N1i—C16—H16B | 109.7 |
| C7—C9—H9A | 109.5 | H16A—C16—H16B | 108.2 |
| C7—C9—H9B | 109.5 | N1—C17—C18 | 109.51 (11) |
| H9A—C9—H9B | 109.5 | N1—C17—H17A | 109.8 |
| C7—C9—H9C | 109.5 | C18—C17—H17A | 109.8 |
| H9A—C9—H9C | 109.5 | N1—C17—H17B | 109.8 |
| H9B—C9—H9C | 109.5 | C18—C17—H17B | 109.8 |
| C7—C10—H10A | 109.5 | H17A—C17—H17B | 108.2 |
| C7—C10—H10B | 109.5 | C17i—C18—C17 | 110.30 (15) |
| H10A—C10—H10B | 109.5 | C17i—C18—H18A | 109.6 |
| C7—C10—H10C | 109.5 | C17—C18—H18A | 109.6 |
| H10A—C10—H10C | 109.5 | C17i—C18—H18B | 109.6 |
| H10B—C10—H10C | 109.5 | C17—C18—H18B | 109.6 |
| C14—C11—C12 | 107.71 (10) | H18A—C18—H18B | 108.1 |
Symmetry code: (i) x, −y+3/2, z.
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O1—H1···N1 | 0.82 | 2.00 | 2.6880 (13) | 142 |
| O2—H2···O1i | 0.85 | 2.22 | 3.036 (5) | 161 |
Symmetry code: (i) x, −y+3/2, z.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: FJ2558).
References
- Barroso, S., Adão, P., Madeira, F., Duarte, M. T., Pessoa, J. C. & Martins, A. M. (2010). Inorg. Chem. 49, 7452–7463. [DOI] [PubMed]
- Bruker (2009). APEX2 and SAINT Bruker AXS Inc., Madison, Wisconsin, USA.
- Guo, Y.-M., Du, M. & Bu, X.-H. (2003). J. Mol. Struct. 646, 191–196.
- Hancock, S. L., Mahon, M. F., Kociok-Köhn, G. & Jones, M. D. (2011). Eur. J. Inorg. Chem. pp. 4596–4602.
- Kannan, S., Kumar, K. N. & Ramesh, R. (2008). Polyhedron, 27, 701–708.
- Manna, C. M., Shavit, M. & Tshuva, E. Y. (2008). J. Organomet. Chem. 693, 3947–3950.
- Mohanty, S., Suresh, D., Balakrishna, M. S. & Mague, J. T. (2008). Tetrahedron, 64, 240–247.
- Pang, M. L., Yao, Y. M., Zhang, Y. & Shen, Q. (2008). Chin. Sci. Bull. 53, 1978–1982.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
- Tshuva, E. Y., Goldberg, I., Kol, M. & Goldschmidt, Z. (2001). Chem. Commun. pp. 2120–2121. [DOI] [PubMed]
- Westrip, S. P. (2010). J. Appl. Cryst. 43, 920–925.
- Wichmann, O., Sillanpää, R. & Lehtonen, A. (2012). Coord. Chem. Rev. 256, 371–392.
- Wong, Y.-L., Tong, L. H., Dilworth, J. R., Ng, D. K. P. & Lee, H. K. (2010). Dalton Trans. 39, 4602–4611. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S1600536812026505/fj2558sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536812026505/fj2558Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report




