Abstract
In the title compound, C17H14O4, the C=C bond adopts an E conformation and the dihedral angle between the benzene rings is 73.9 (1)°. The crystal packing features C—H⋯O hydrogen bonds, which generate C(4) chains propagating along the b-axis direction. Weak aromatic π–π stacking interactions [centroid–centroid distance = 3.703 (1) Å] are also observed.
Related literature
For the biological properties of cinnamate derivatives, see: Sharma (2011 ▶). For related structures, see: Kaitner & Stilinović (2007 ▶); Anuradha et al. (2012 ▶).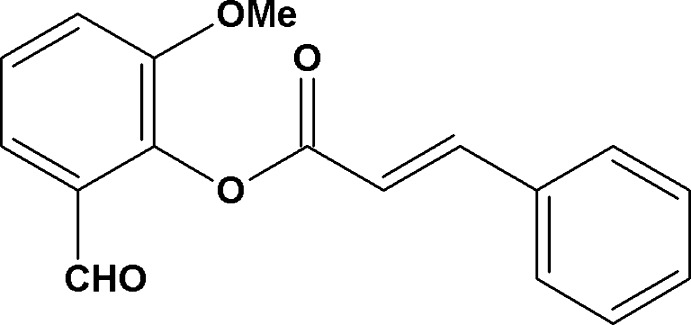
Experimental
Crystal data
C17H14O4
M r = 282.28
Orthorhombic,

a = 10.7908 (7) Å
b = 10.4672 (5) Å
c = 25.8714 (17) Å
V = 2922.2 (3) Å3
Z = 8
Mo Kα radiation
μ = 0.09 mm−1
T = 293 K
0.23 × 0.21 × 0.15 mm
Data collection
Bruker APEXII CCD diffractometer
Absorption correction: multi-scan (SADABS; Sheldrick, 1996 ▶) T min = 0.910, T max = 0.941
14344 measured reflections
2665 independent reflections
1909 reflections with I > 2σ(I)
R int = 0.026
Refinement
R[F 2 > 2σ(F 2)] = 0.037
wR(F 2) = 0.106
S = 1.01
2665 reflections
192 parameters
H-atom parameters constrained
Δρmax = 0.15 e Å−3
Δρmin = −0.12 e Å−3
Data collection: APEX2 (Bruker, 2004 ▶); cell refinement: APEX2 and SAINT (Bruker, 2004 ▶); data reduction: SAINT and XPREP (Bruker, 2004 ▶); program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: ORTEP-3 (Farrugia (1997 ▶); software used to prepare material for publication: SHELXL97 and PLATON (Spek, 2009 ▶).
Supplementary Material
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S160053681202747X/hb6858sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S160053681202747X/hb6858Isup2.hkl
Supplementary material file. DOI: 10.1107/S160053681202747X/hb6858Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C9—H9⋯O3i | 0.93 | 2.50 | 3.415 (2) | 168 |
Symmetry code: (i)  .
.
Acknowledgments
The authors thank Dr Babu Vargheese, SAIF, IIT, Madras, India, for his help with the data collection.
supplementary crystallographic information
Comment
Cinnamic acid and its derivatives including esters and carboxylic functional derivatives are used as important components in flavours, perfumes, synthetic indigo and pharmaceuticals. Cinnamate can act as optical filters or deactivate substrate molecules that have been excited by light for the protection polymers and organic substances. They are used as cosmetic grades and as sunscreen agents to reduce skin damage by blocking UV—A, B (Sharma, 2011). As part of our studies in this area, the crystal structure determination of the title compound was carried out and the results are presented here.
The C9=C10 double bond of the title compound (I) exists in an E-configuration (Fig. 1). The dihedral angle between the two benzene rings is 73.9 (1)°. The geometric parameters of the title molecule agrees well with those reported for similar structures (Kaitner et al., 2007, Anuradha et al., 2012).
The crystal packing features C—H···O hydrogen bonds. Atom C9 at x, y, z donates one proton to atom O3 at 3/2 - x, 1/2 + y, z, forming C(4) chains along the b axis (Fig. 2). The crystal packing (Fig. 3) also features π—π interactions with a Cg—Cgiv seperation of 3.703 (1) Å.[Fig. 3; Cg is the centroid of the C1–C6 benzene ring, symmetry code as in Fig. 3].
Experimental
To a stirred solution of 2-hydroxy-3-methoxybenzaldehyde (1 mmol, 0.154 g) and potassium carbonate (1.5 mmol, 0.207 g) was stirred for 15 minutes in acetonitrile as solvent at room temperature. To this solution, (2E)-3-phenylprop-2-enoylchloride (1 mmol, 0.167 g) was added till the addition is complete. After the completion of the reaction as indicated by TLC, acetonitrile solvent was evaporated. Ethylacetate (15 ml) and water (15 ml) were added to the crude mass. The organic layer was dried over anhydrous sodium sulfate. Removal of solvent led to the crude product. The pure title compound was obtained as a colorless solid (0.250 g, 89% yield). Recrystallization was carried out using ethylacetate as solvent to yield colourless blocks.
Refinement
H atoms were positioned geometrically, with C–H = 0.93–0.96 Å and constrained to ride on their parent atoms, with Uiso(H) =1.5Ueq for methyl H atoms and 1.2Ueq(C) for other H atoms.
Figures
Fig. 1.
Molecular structure of the title compound showing displacement ellipsoids at the 30% probability level.
Fig. 2.
Part of the crystal structure of (I) showing intermolecular C—H···O hydrogen bonds (dotted lines), forming C(4) chains along the b axis. For clarity H atoms involved in the hydrogen bonds are shown. [Symmetry codes:(i)3/2 - x, 1/2 + y, z; (ii)x, 1 + y, z; (iii)3/2 - x, 3/2 + y, z].
Fig. 3.
A view of the π—π interactions (dotted lines) in the crystal structure of the title compound. Cg denotes centroid of the C1–C6 benzene ring. [Symmetry code: (iv)1 - x, -y, 1 - z].
Crystal data
| C17H14O4 | F(000) = 1184 |
| Mr = 282.28 | Dx = 1.283 Mg m−3 |
| Orthorhombic, Pbca | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2ac 2ab | Cell parameters from 2879 reflections |
| a = 10.7908 (7) Å | θ = 1.0–1.0° |
| b = 10.4672 (5) Å | µ = 0.09 mm−1 |
| c = 25.8714 (17) Å | T = 293 K |
| V = 2922.2 (3) Å3 | Block, colourless |
| Z = 8 | 0.23 × 0.21 × 0.15 mm |
Data collection
| Bruker APEXII CCD diffractometer | 2665 independent reflections |
| Radiation source: fine-focus sealed tube | 1909 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.026 |
| Detector resolution: 10.0 pixels mm-1 | θmax = 26.0°, θmin = 2.5° |
| ω scans | h = −13→13 |
| Absorption correction: multi-scan (SADABS; Sheldrick, 1996) | k = −9→11 |
| Tmin = 0.910, Tmax = 0.941 | l = −31→31 |
| 14344 measured reflections |
Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.037 | H-atom parameters constrained |
| wR(F2) = 0.106 | w = 1/[σ2(Fo2) + (0.0446P)2 + 0.7231P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.01 | (Δ/σ)max = 0.001 |
| 2665 reflections | Δρmax = 0.15 e Å−3 |
| 192 parameters | Δρmin = −0.12 e Å−3 |
| 0 restraints | Extinction correction: SHELXL97 (Sheldrick, 2008), Fc*=kFc[1+0.001xFc2λ3/sin(2θ)]-1/4 |
| Primary atom site location: structure-invariant direct methods | Extinction coefficient: 0.0020 (5) |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| C1 | 0.62871 (15) | −0.06915 (15) | 0.54858 (6) | 0.0487 (4) | |
| C2 | 0.54050 (16) | −0.15242 (16) | 0.56771 (7) | 0.0552 (4) | |
| C3 | 0.48566 (18) | −0.23727 (18) | 0.53357 (8) | 0.0686 (5) | |
| H3 | 0.4252 | −0.2935 | 0.5453 | 0.082* | |
| C4 | 0.5205 (2) | −0.2388 (2) | 0.48191 (8) | 0.0732 (6) | |
| H4 | 0.4827 | −0.2959 | 0.4593 | 0.088* | |
| C5 | 0.60927 (19) | −0.15776 (18) | 0.46393 (7) | 0.0649 (5) | |
| H5 | 0.6326 | −0.1603 | 0.4293 | 0.078* | |
| C6 | 0.66465 (16) | −0.07149 (16) | 0.49726 (7) | 0.0532 (4) | |
| C7 | 0.7586 (2) | 0.0175 (2) | 0.47767 (8) | 0.0702 (5) | |
| H7 | 0.7865 | 0.0813 | 0.4998 | 0.084* | |
| C8 | 0.76334 (15) | −0.01222 (16) | 0.61648 (6) | 0.0459 (4) | |
| C9 | 0.78497 (15) | 0.08922 (16) | 0.65399 (6) | 0.0491 (4) | |
| H9 | 0.7521 | 0.1700 | 0.6480 | 0.059* | |
| C10 | 0.85036 (15) | 0.06857 (17) | 0.69633 (6) | 0.0509 (4) | |
| H10 | 0.8863 | −0.0119 | 0.6993 | 0.061* | |
| C11 | 0.87318 (14) | 0.15626 (16) | 0.73902 (6) | 0.0484 (4) | |
| C12 | 0.82650 (17) | 0.28000 (17) | 0.73974 (6) | 0.0595 (5) | |
| H12 | 0.7812 | 0.3101 | 0.7117 | 0.071* | |
| C13 | 0.84677 (18) | 0.35820 (19) | 0.78160 (7) | 0.0663 (5) | |
| H13 | 0.8149 | 0.4407 | 0.7816 | 0.080* | |
| C14 | 0.91335 (17) | 0.3161 (2) | 0.82338 (7) | 0.0667 (5) | |
| H14 | 0.9258 | 0.3694 | 0.8517 | 0.080* | |
| C15 | 0.96137 (18) | 0.1953 (2) | 0.82313 (7) | 0.0703 (6) | |
| H15 | 1.0074 | 0.1665 | 0.8512 | 0.084* | |
| C16 | 0.94166 (17) | 0.11608 (19) | 0.78136 (6) | 0.0619 (5) | |
| H16 | 0.9749 | 0.0341 | 0.7816 | 0.074* | |
| C17 | 0.41618 (19) | −0.2177 (2) | 0.63873 (9) | 0.0923 (7) | |
| H17A | 0.3401 | −0.1947 | 0.6219 | 0.138* | |
| H17B | 0.4085 | −0.2032 | 0.6752 | 0.138* | |
| H17C | 0.4336 | −0.3063 | 0.6325 | 0.138* | |
| O1 | 0.80202 (17) | 0.01350 (16) | 0.43495 (6) | 0.0987 (5) | |
| O2 | 0.67581 (11) | 0.02494 (10) | 0.58150 (4) | 0.0542 (3) | |
| O3 | 0.81108 (12) | −0.11529 (12) | 0.61497 (5) | 0.0630 (4) | |
| O4 | 0.51428 (13) | −0.14196 (13) | 0.61881 (5) | 0.0731 (4) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| C1 | 0.0519 (10) | 0.0386 (10) | 0.0557 (9) | 0.0070 (7) | −0.0156 (8) | −0.0053 (7) |
| C2 | 0.0530 (10) | 0.0508 (11) | 0.0617 (10) | 0.0072 (8) | −0.0079 (8) | −0.0057 (8) |
| C3 | 0.0582 (11) | 0.0561 (12) | 0.0914 (15) | −0.0041 (9) | −0.0122 (10) | −0.0078 (10) |
| C4 | 0.0754 (13) | 0.0635 (14) | 0.0806 (14) | 0.0058 (11) | −0.0266 (11) | −0.0246 (11) |
| C5 | 0.0766 (13) | 0.0622 (13) | 0.0560 (10) | 0.0125 (11) | −0.0150 (10) | −0.0123 (9) |
| C6 | 0.0599 (11) | 0.0471 (11) | 0.0527 (9) | 0.0099 (8) | −0.0109 (8) | −0.0010 (8) |
| C7 | 0.0815 (14) | 0.0650 (14) | 0.0641 (11) | 0.0035 (11) | −0.0039 (11) | 0.0060 (10) |
| C8 | 0.0482 (9) | 0.0427 (11) | 0.0468 (8) | −0.0014 (8) | −0.0038 (7) | 0.0034 (7) |
| C9 | 0.0550 (10) | 0.0404 (10) | 0.0520 (9) | −0.0011 (8) | −0.0064 (8) | −0.0009 (7) |
| C10 | 0.0528 (10) | 0.0467 (11) | 0.0531 (9) | 0.0010 (8) | −0.0034 (8) | 0.0018 (7) |
| C11 | 0.0483 (9) | 0.0520 (11) | 0.0449 (8) | −0.0040 (8) | −0.0014 (7) | 0.0005 (7) |
| C12 | 0.0710 (12) | 0.0564 (12) | 0.0510 (10) | −0.0017 (9) | −0.0106 (9) | −0.0022 (8) |
| C13 | 0.0793 (13) | 0.0588 (12) | 0.0607 (11) | −0.0032 (10) | −0.0011 (10) | −0.0107 (9) |
| C14 | 0.0587 (11) | 0.0911 (16) | 0.0502 (10) | −0.0126 (11) | 0.0015 (9) | −0.0172 (10) |
| C15 | 0.0601 (11) | 0.1040 (17) | 0.0469 (9) | 0.0032 (11) | −0.0090 (8) | −0.0033 (10) |
| C16 | 0.0625 (11) | 0.0700 (13) | 0.0532 (10) | 0.0090 (9) | −0.0073 (9) | 0.0018 (9) |
| C17 | 0.0650 (13) | 0.118 (2) | 0.0936 (16) | −0.0043 (13) | 0.0096 (12) | 0.0217 (14) |
| O1 | 0.1247 (14) | 0.0973 (12) | 0.0741 (10) | −0.0031 (10) | 0.0230 (9) | 0.0072 (8) |
| O2 | 0.0664 (7) | 0.0407 (7) | 0.0556 (6) | 0.0052 (5) | −0.0191 (6) | −0.0055 (5) |
| O3 | 0.0680 (8) | 0.0495 (8) | 0.0717 (8) | 0.0140 (6) | −0.0187 (6) | −0.0094 (6) |
| O4 | 0.0713 (9) | 0.0803 (10) | 0.0678 (8) | −0.0064 (7) | 0.0058 (7) | −0.0038 (7) |
Geometric parameters (Å, º)
| C1—C2 | 1.382 (2) | C9—H9 | 0.9300 |
| C1—C6 | 1.383 (2) | C10—C11 | 1.457 (2) |
| C1—O2 | 1.3978 (19) | C10—H10 | 0.9300 |
| C2—O4 | 1.356 (2) | C11—C16 | 1.387 (2) |
| C2—C3 | 1.385 (2) | C11—C12 | 1.390 (2) |
| C3—C4 | 1.389 (3) | C12—C13 | 1.375 (2) |
| C3—H3 | 0.9300 | C12—H12 | 0.9300 |
| C4—C5 | 1.361 (3) | C13—C14 | 1.371 (3) |
| C4—H4 | 0.9300 | C13—H13 | 0.9300 |
| C5—C6 | 1.384 (2) | C14—C15 | 1.366 (3) |
| C5—H5 | 0.9300 | C14—H14 | 0.9300 |
| C6—C7 | 1.468 (3) | C15—C16 | 1.379 (3) |
| C7—O1 | 1.201 (2) | C15—H15 | 0.9300 |
| C7—H7 | 0.9300 | C16—H16 | 0.9300 |
| C8—O3 | 1.1961 (19) | C17—O4 | 1.419 (2) |
| C8—O2 | 1.3646 (18) | C17—H17A | 0.9600 |
| C8—C9 | 1.457 (2) | C17—H17B | 0.9600 |
| C9—C10 | 1.321 (2) | C17—H17C | 0.9600 |
| C2—C1—C6 | 121.71 (15) | C9—C10—H10 | 116.0 |
| C2—C1—O2 | 118.45 (15) | C11—C10—H10 | 116.0 |
| C6—C1—O2 | 119.70 (15) | C16—C11—C12 | 117.72 (15) |
| O4—C2—C1 | 116.23 (15) | C16—C11—C10 | 119.85 (16) |
| O4—C2—C3 | 125.74 (18) | C12—C11—C10 | 122.41 (15) |
| C1—C2—C3 | 118.03 (17) | C13—C12—C11 | 120.51 (17) |
| C2—C3—C4 | 120.37 (19) | C13—C12—H12 | 119.7 |
| C2—C3—H3 | 119.8 | C11—C12—H12 | 119.7 |
| C4—C3—H3 | 119.8 | C14—C13—C12 | 120.87 (19) |
| C5—C4—C3 | 120.80 (18) | C14—C13—H13 | 119.6 |
| C5—C4—H4 | 119.6 | C12—C13—H13 | 119.6 |
| C3—C4—H4 | 119.6 | C15—C14—C13 | 119.51 (18) |
| C4—C5—C6 | 119.83 (18) | C15—C14—H14 | 120.2 |
| C4—C5—H5 | 120.1 | C13—C14—H14 | 120.2 |
| C6—C5—H5 | 120.1 | C14—C15—C16 | 120.12 (18) |
| C1—C6—C5 | 119.24 (18) | C14—C15—H15 | 119.9 |
| C1—C6—C7 | 120.93 (16) | C16—C15—H15 | 119.9 |
| C5—C6—C7 | 119.82 (17) | C15—C16—C11 | 121.26 (18) |
| O1—C7—C6 | 124.4 (2) | C15—C16—H16 | 119.4 |
| O1—C7—H7 | 117.8 | C11—C16—H16 | 119.4 |
| C6—C7—H7 | 117.8 | O4—C17—H17A | 109.5 |
| O3—C8—O2 | 122.24 (14) | O4—C17—H17B | 109.5 |
| O3—C8—C9 | 127.58 (15) | H17A—C17—H17B | 109.5 |
| O2—C8—C9 | 110.17 (14) | O4—C17—H17C | 109.5 |
| C10—C9—C8 | 121.24 (16) | H17A—C17—H17C | 109.5 |
| C10—C9—H9 | 119.4 | H17B—C17—H17C | 109.5 |
| C8—C9—H9 | 119.4 | C8—O2—C1 | 117.06 (12) |
| C9—C10—C11 | 128.01 (16) | C2—O4—C17 | 117.68 (16) |
| C6—C1—C2—O4 | 179.15 (15) | C8—C9—C10—C11 | −175.41 (15) |
| O2—C1—C2—O4 | −5.3 (2) | C9—C10—C11—C16 | 178.04 (17) |
| C6—C1—C2—C3 | −1.7 (2) | C9—C10—C11—C12 | −0.6 (3) |
| O2—C1—C2—C3 | 173.89 (14) | C16—C11—C12—C13 | −0.9 (3) |
| O4—C2—C3—C4 | 179.98 (17) | C10—C11—C12—C13 | 177.80 (17) |
| C1—C2—C3—C4 | 0.9 (3) | C11—C12—C13—C14 | 0.1 (3) |
| C2—C3—C4—C5 | 0.3 (3) | C12—C13—C14—C15 | 0.8 (3) |
| C3—C4—C5—C6 | −0.8 (3) | C13—C14—C15—C16 | −0.7 (3) |
| C2—C1—C6—C5 | 1.2 (2) | C14—C15—C16—C11 | −0.1 (3) |
| O2—C1—C6—C5 | −174.30 (14) | C12—C11—C16—C15 | 0.9 (3) |
| C2—C1—C6—C7 | −179.88 (16) | C10—C11—C16—C15 | −177.80 (17) |
| O2—C1—C6—C7 | 4.6 (2) | O3—C8—O2—C1 | 10.1 (2) |
| C4—C5—C6—C1 | 0.0 (3) | C9—C8—O2—C1 | −169.00 (14) |
| C4—C5—C6—C7 | −178.85 (17) | C2—C1—O2—C8 | 78.41 (18) |
| C1—C6—C7—O1 | 173.14 (19) | C6—C1—O2—C8 | −105.90 (17) |
| C5—C6—C7—O1 | −8.0 (3) | C1—C2—O4—C17 | 174.31 (16) |
| O3—C8—C9—C10 | −10.2 (3) | C3—C2—O4—C17 | −4.8 (3) |
| O2—C8—C9—C10 | 168.89 (15) |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| C9—H9···O3i | 0.93 | 2.50 | 3.415 (2) | 168 |
Symmetry code: (i) −x+3/2, y+1/2, z.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: HB6858).
References
- Anuradha, T., Sivakumar, G., Seshadri, P. R. & Bakthadoss, M. (2012). Acta Cryst. E68, o229. [DOI] [PMC free article] [PubMed]
- Bruker (2004). APEX2, SAINT and XPREP Bruker AXS Inc., Madison, Wisconsin, USA.
- Farrugia, L. J. (1997). J. Appl. Cryst. 30, 565.
- Kaitner, B. & Stilinović, V. (2007). Acta Cryst. E63, o4347. [DOI] [PubMed]
- Sharma, P. (2011). J. Chem. Pharm. Res. 3, 403–423.
- Sheldrick, G. M. (1996). SADABS University of Göttingen, Germany.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S160053681202747X/hb6858sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S160053681202747X/hb6858Isup2.hkl
Supplementary material file. DOI: 10.1107/S160053681202747X/hb6858Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report





