Abstract
The crystal structure of the title compound, C15H14N4, contains chains of coplanar tetrazole rings with the chain direction along b. These are formed through weak hydrogen bonds, donated by the tetrazole H atoms and by one of the H atoms of the methylene group, and accepted by two neighbouring N atoms of the adjacent tetrazole ring. The chains are connected to each other in a staircase-like manner via weak hydrogen bonds, donated from the second H atom of the methylene group and accepted by the N atom next to the C atom in the tetrazole ring. The resulting layers are parallel to the bc plane.
Related literature
For the synthesis, see Kamiya & Saito (1973 ▶). For crystal structure studies of 1H-tetrazol-1-yl compounds, see Absmeier et al. (2006 ▶); Grunert et al. (2005 ▶); Werner et al. (2009 ▶).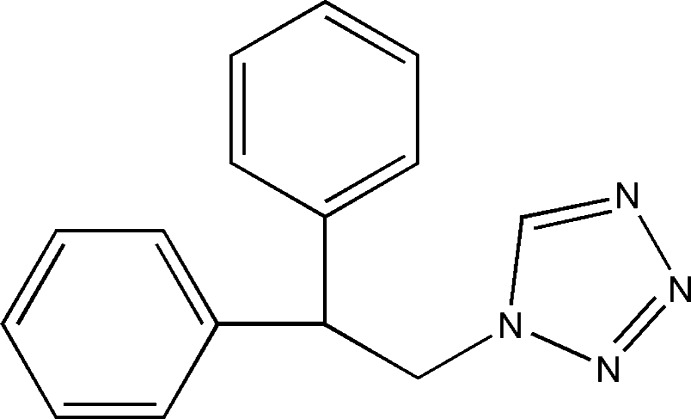
Experimental
Crystal data
C15H14N4
M r = 250.30
Monoclinic,

a = 12.5289 (6) Å
b = 10.4157 (5) Å
c = 11.0085 (5) Å
β = 107.906 (1)°
V = 1366.99 (11) Å3
Z = 4
Mo Kα radiation
μ = 0.08 mm−1
T = 296 K
0.45 × 0.40 × 0.35 mm
Data collection
Bruker APEXII CCD diffractometer
Absorption correction: multi-scan (SADABS; Sheldrick, 2008b ▶) T min = 0.89, T max = 0.97
18385 measured reflections
3984 independent reflections
3230 reflections with I > 2σ(I)
R int = 0.019
Refinement
R[F 2 > 2σ(F 2)] = 0.050
wR(F 2) = 0.148
S = 1.04
3984 reflections
172 parameters
H-atom parameters constrained
Δρmax = 0.22 e Å−3
Δρmin = −0.16 e Å−3
Data collection: APEX2 (Bruker, 2011 ▶); cell refinement: SAINT (Bruker, 2009 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008a ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008a ▶); molecular graphics: ATOMS (Dowty, 2006 ▶), Mercury (Macrae et al., 2006 ▶) and VESTA (Momma & Izumi, 2008 ▶); software used to prepare material for publication: SHELXL97.
Supplementary Material
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536812027675/fj2563sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536812027675/fj2563Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536812027675/fj2563Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C1—H1⋯N3i | 0.93 | 2.61 | 3.4690 (19) | 153 |
| C2—H2B⋯N4i | 0.97 | 2.50 | 3.4622 (17) | 174 |
| C2—H2A⋯N4ii | 0.97 | 2.56 | 3.5155 (17) | 169 |
Symmetry codes: (i)  ; (ii)
; (ii)  .
.
Acknowledgments
Thanks for financial support are due to the "Fonds zur Förderung der Wissenschaftlichen Forschung in Österreich" (project 19335-N17).
supplementary crystallographic information
Comment
In continuation of the crystallographic characterization of 1H–tetrazol–1–yl compounds, intended as potential ligands for Fe(II) spin crossover complexes (Absmeier et al., 2006; Grunert et al., 2005; Werner et al., 2009), the title compound was prepared.
At 296 K the title compound crystallizes in the monoclinic space group P21/c (No. 14), with one molecule in the asymmetric unit (Fig. 1). Bond lenghts and bond angles in the molecule adopt typical values. The point group symmetry of the free molecule is Cs. Owing to intermolecular interactions this symmetry is lowered to C1 in the crystalline solid, which can be readily seen from the (+)-synclinal arrangement of the tetrazolyl ring and the phenyl ring C10···C15 [N1—C2—C3—C10 = 60.68 (12)°] and the out–of–plane (plane defined by N1, C2 and C3) twist of the tetrazolyl ring [N2—N1—C2—C3 = 63.98 (14)°].
In the crystal the main type of interaction are three sets of weak hydrogen bonds between the tetrazolyl rings (C1—H1···N3) and the tetrazolyl rings and the methylenic H atoms (C2—H2A···N4 and C2—H2B···N4). Through the coplanar interactions C1—H1···N3 and C2—H2B···N4 chains of tetrazolyl rings are formed parallel to the b-axis (Fig. 2), whereas C2—H2A···N4 connects these chains in a staircase-like manner (Fig. 3) resulting in the formation of layers parallel to the bc-plane with the phenyl rings pointing outwards. The layers are loosely held together by C—H···π interactions (Fig. 4).
Experimental
The title compound was prepared according to the general procedure given by Kamiya & Saito (1973). All chemicals were used as supplied without further purification. Elemental analyses were performed on a Perkin Elmer 2400 CHN Elemental Analyzer. NMR-spectra were measured in DMSO-d6 with a Bruker DPX-200 spectrometer at 200 MHz (1H) and 50 MHz (13C) respectively. The chemical shifts (see Fig. 5 for the atom assignment) are calibrated to the solvent.
2,2-Diphenylethylamine (4.93 g, 25 mmol, Aldrich, 96%), NaN3 (3.25 g, 50 mmol, Fluka, 99%) and triethyl orthoformate (7.42 g, 50 mmol, Acros, 98%) were dissolved in 60 ml of acetic acid (Fluka, 99.8%) and heated to 85–90°C for 24 h. After evaporation of acetic acid under reduced pressure, 70 ml of 2 N hydrochloric acid was added to the residue. The solution was extracted three times with dichloromethane, the combined organic layers were washed with water and saturated aqueous solutions of NaHCO3 and NaCl. The organic phase was dried with Na2SO4, filtered and the solvent was evaporated under reduced pressure. The raw product was recrystallized from methanol. Yield 2.88 g (46%), m.p. 115°C. Single crystals were grown by slow evaporation from a solution of the tetrazole in methanol at room temperature over two days.
Elemental analysis C15H14N4 Calc.: C 71.98, H 5.64, N 22.38. Found: C 72.10, H 5.38, N 21.91%. 1H-NMR (DMSO-d6) δ [p.p.m.]: 4.73 (t, 3J=8.4 Hz, 1H, Hc), 5.21 (d, 3J=8.5 Hz, 2H, Hb), 7.15, 7.19, 7.22, 7.25, 7.29, 7.32, 7.36, 7.40 (m, 10H, He–g), 9.24 (s, 1H, Ha). 13C-NMR (DMSO-d6) δ [p.p.m.]: 50.5, 50.8 (Cb–c), 127.0 (Cg), 127.8 (Ce), 128.7 (Cf), 140.7 (Cd), 144.0 (Ca).
Refinement
Hydrogen atoms were included at calculated positions and treated as riding on their base atoms with d(C—H)= 0.97 (CH2), 0.98 (CH) or 0.93 Å (CHarom) and Uiso(H)=1.2Ueq(C). Reflection 020 was omitted because of its large Δ(F2)/e.s.d. value.
Figures
Fig. 1.
Molecular moiety in the crystal structure of the title compound. Displacement ellipsoids for non–H atoms are drawn at the 33% probability level. Hydrogen atoms involved in weak hydrogen bonding are labelled.
Fig. 2.
Weak hydrogen bonds in the crystal structure of the title compound, viewed perpedicular to the layers formed by adjacent tetrazolyl rings (yellow C1—H1···N3, red C2—H2B···N4, orange C2—H2A···N4; phenyl rings are omitted for clarity).
Fig. 3.
View of the weak hydrogen bonds in the crystal structure of the title compound, showing the staircase-like arrangement (yellow C1—H1···N3, red C2—H2B···N4, orange C2—H2A···N4; phenyl rings are omitted for clarity).
Fig. 4.
Packing diagram of the title compound. Weak hydrogen bonds are drawn with cyan dashed lines, only those hydrogen atoms involved in weak interactions are shown and the unit cell is outlined.
Fig. 5.
Labelling scheme for the assignment of the NMR chemical shifts.
Crystal data
| C15H14N4 | F(000) = 528 |
| Mr = 250.30 | Dx = 1.216 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2ybc | Cell parameters from 8429 reflections |
| a = 12.5289 (6) Å | θ = 2.6–30.0° |
| b = 10.4157 (5) Å | µ = 0.08 mm−1 |
| c = 11.0085 (5) Å | T = 296 K |
| β = 107.906 (1)° | Block, colourless |
| V = 1366.99 (11) Å3 | 0.45 × 0.40 × 0.35 mm |
| Z = 4 |
Data collection
| Bruker APEXII CCD diffractometer | 3984 independent reflections |
| Radiation source: fine-focus sealed tube | 3230 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.019 |
| φ and ω scans | θmax = 30.1°, θmin = 3.4° |
| Absorption correction: multi-scan (SADABS; Sheldrick, 2008b) | h = −17→17 |
| Tmin = 0.89, Tmax = 0.97 | k = −14→14 |
| 18385 measured reflections | l = −15→15 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.050 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.148 | H-atom parameters constrained |
| S = 1.04 | w = 1/[σ2(Fo2) + (0.0655P)2 + 0.2149P] where P = (Fo2 + 2Fc2)/3 |
| 3984 reflections | (Δ/σ)max < 0.001 |
| 172 parameters | Δρmax = 0.22 e Å−3 |
| 0 restraints | Δρmin = −0.16 e Å−3 |
Special details
| Experimental. Bruker Kappa APEX2 CCD diffractometer, full-sphere data collection. |
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| N1 | 0.06288 (8) | 0.25473 (9) | 0.39090 (9) | 0.0503 (2) | |
| N2 | 0.07582 (12) | 0.13614 (12) | 0.43964 (12) | 0.0755 (4) | |
| N3 | 0.03463 (12) | 0.05898 (12) | 0.34413 (14) | 0.0804 (4) | |
| N4 | −0.00552 (11) | 0.12529 (13) | 0.23470 (12) | 0.0724 (3) | |
| C1 | 0.01312 (12) | 0.24583 (14) | 0.26661 (13) | 0.0630 (3) | |
| H1 | −0.0057 | 0.3149 | 0.2105 | 0.076* | |
| C2 | 0.09677 (9) | 0.36869 (11) | 0.47103 (11) | 0.0493 (2) | |
| H2A | 0.0581 | 0.3693 | 0.5351 | 0.059* | |
| H2B | 0.0740 | 0.4448 | 0.4186 | 0.059* | |
| C3 | 0.22336 (9) | 0.37415 (10) | 0.53767 (10) | 0.0464 (2) | |
| H3 | 0.2443 | 0.2965 | 0.5899 | 0.056* | |
| C4 | 0.24642 (9) | 0.48874 (12) | 0.62769 (11) | 0.0506 (3) | |
| C5 | 0.29138 (12) | 0.47133 (17) | 0.75794 (13) | 0.0707 (4) | |
| H5 | 0.3081 | 0.3892 | 0.7915 | 0.085* | |
| C6 | 0.31156 (15) | 0.5784 (2) | 0.83910 (16) | 0.0925 (6) | |
| H6 | 0.3422 | 0.5670 | 0.9268 | 0.111* | |
| C7 | 0.28658 (15) | 0.6999 (2) | 0.7905 (2) | 0.0903 (6) | |
| H7 | 0.3007 | 0.7703 | 0.8451 | 0.108* | |
| C8 | 0.24121 (13) | 0.71720 (16) | 0.66276 (19) | 0.0804 (5) | |
| H8 | 0.2235 | 0.7995 | 0.6300 | 0.096* | |
| C9 | 0.22120 (11) | 0.61269 (12) | 0.58099 (14) | 0.0617 (3) | |
| H9 | 0.1904 | 0.6257 | 0.4936 | 0.074* | |
| C10 | 0.29411 (8) | 0.37823 (10) | 0.44771 (10) | 0.0458 (2) | |
| C11 | 0.40353 (11) | 0.33206 (17) | 0.49185 (14) | 0.0722 (4) | |
| H11 | 0.4298 | 0.2966 | 0.5732 | 0.087* | |
| C12 | 0.47312 (12) | 0.3379 (2) | 0.41749 (19) | 0.0919 (6) | |
| H12 | 0.5460 | 0.3068 | 0.4490 | 0.110* | |
| C13 | 0.43670 (13) | 0.38890 (17) | 0.29785 (18) | 0.0805 (5) | |
| H13 | 0.4848 | 0.3937 | 0.2483 | 0.097* | |
| C14 | 0.32823 (14) | 0.43332 (14) | 0.25067 (15) | 0.0701 (4) | |
| H14 | 0.3024 | 0.4666 | 0.1684 | 0.084* | |
| C15 | 0.25730 (11) | 0.42836 (12) | 0.32603 (12) | 0.0563 (3) | |
| H15 | 0.1843 | 0.4592 | 0.2940 | 0.068* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| N1 | 0.0485 (5) | 0.0525 (5) | 0.0533 (5) | −0.0114 (4) | 0.0206 (4) | −0.0081 (4) |
| N2 | 0.0969 (9) | 0.0575 (6) | 0.0699 (7) | −0.0239 (6) | 0.0225 (6) | −0.0022 (5) |
| N3 | 0.0952 (9) | 0.0597 (7) | 0.0895 (9) | −0.0277 (6) | 0.0329 (7) | −0.0172 (6) |
| N4 | 0.0727 (7) | 0.0758 (8) | 0.0703 (7) | −0.0213 (6) | 0.0242 (6) | −0.0241 (6) |
| C1 | 0.0632 (7) | 0.0674 (8) | 0.0556 (7) | −0.0093 (6) | 0.0141 (5) | −0.0115 (6) |
| C2 | 0.0484 (5) | 0.0522 (6) | 0.0515 (6) | −0.0071 (4) | 0.0214 (4) | −0.0102 (4) |
| C3 | 0.0499 (5) | 0.0438 (5) | 0.0453 (5) | −0.0031 (4) | 0.0146 (4) | −0.0017 (4) |
| C4 | 0.0465 (5) | 0.0573 (6) | 0.0506 (5) | −0.0084 (4) | 0.0189 (4) | −0.0114 (5) |
| C5 | 0.0712 (8) | 0.0886 (10) | 0.0520 (7) | −0.0096 (7) | 0.0186 (6) | −0.0095 (7) |
| C6 | 0.0856 (11) | 0.1339 (18) | 0.0576 (8) | −0.0170 (11) | 0.0215 (7) | −0.0333 (10) |
| C7 | 0.0762 (9) | 0.0991 (13) | 0.0996 (13) | −0.0139 (9) | 0.0326 (9) | −0.0541 (11) |
| C8 | 0.0698 (8) | 0.0656 (8) | 0.1080 (13) | −0.0083 (7) | 0.0306 (8) | −0.0332 (8) |
| C9 | 0.0611 (7) | 0.0550 (7) | 0.0701 (8) | −0.0065 (5) | 0.0217 (6) | −0.0131 (6) |
| C10 | 0.0442 (5) | 0.0420 (5) | 0.0521 (5) | −0.0029 (4) | 0.0164 (4) | −0.0079 (4) |
| C11 | 0.0502 (6) | 0.0951 (11) | 0.0667 (8) | 0.0103 (6) | 0.0110 (6) | −0.0048 (7) |
| C12 | 0.0472 (7) | 0.1337 (16) | 0.0969 (12) | 0.0088 (8) | 0.0253 (7) | −0.0167 (11) |
| C13 | 0.0695 (8) | 0.0884 (11) | 0.1028 (12) | −0.0155 (7) | 0.0549 (9) | −0.0259 (9) |
| C14 | 0.0884 (10) | 0.0626 (8) | 0.0741 (8) | −0.0027 (7) | 0.0470 (7) | 0.0009 (6) |
| C15 | 0.0588 (6) | 0.0527 (6) | 0.0641 (7) | 0.0081 (5) | 0.0285 (5) | 0.0063 (5) |
Geometric parameters (Å, º)
| N1—C1 | 1.3209 (16) | C6—H6 | 0.9300 |
| N1—N2 | 1.3365 (15) | C7—C8 | 1.357 (3) |
| N1—C2 | 1.4619 (13) | C7—H7 | 0.9300 |
| N2—N3 | 1.2978 (17) | C8—C9 | 1.3854 (18) |
| N3—N4 | 1.3449 (19) | C8—H8 | 0.9300 |
| N4—C1 | 1.3054 (17) | C9—H9 | 0.9300 |
| C1—H1 | 0.9300 | C10—C15 | 1.3783 (17) |
| C2—C3 | 1.5301 (15) | C10—C11 | 1.3918 (16) |
| C2—H2A | 0.9700 | C11—C12 | 1.368 (2) |
| C2—H2B | 0.9700 | C11—H11 | 0.9300 |
| C3—C10 | 1.5193 (14) | C12—C13 | 1.362 (3) |
| C3—C4 | 1.5212 (15) | C12—H12 | 0.9300 |
| C3—H3 | 0.9800 | C13—C14 | 1.377 (2) |
| C4—C5 | 1.3818 (18) | C13—H13 | 0.9300 |
| C4—C9 | 1.3896 (18) | C14—C15 | 1.3906 (17) |
| C5—C6 | 1.403 (2) | C14—H14 | 0.9300 |
| C5—H5 | 0.9300 | C15—H15 | 0.9300 |
| C6—C7 | 1.372 (3) | ||
| C1—N1—N2 | 108.14 (11) | C5—C6—H6 | 119.7 |
| C1—N1—C2 | 129.72 (11) | C8—C7—C6 | 119.97 (15) |
| N2—N1—C2 | 122.09 (10) | C8—C7—H7 | 120.0 |
| N3—N2—N1 | 106.15 (12) | C6—C7—H7 | 120.0 |
| N2—N3—N4 | 110.72 (12) | C7—C8—C9 | 120.21 (17) |
| C1—N4—N3 | 105.44 (11) | C7—C8—H8 | 119.9 |
| N4—C1—N1 | 109.55 (13) | C9—C8—H8 | 119.9 |
| N4—C1—H1 | 125.2 | C8—C9—C4 | 120.96 (14) |
| N1—C1—H1 | 125.2 | C8—C9—H9 | 119.5 |
| N1—C2—C3 | 112.67 (9) | C4—C9—H9 | 119.5 |
| N1—C2—H2A | 109.1 | C15—C10—C11 | 118.00 (11) |
| C3—C2—H2A | 109.1 | C15—C10—C3 | 123.79 (10) |
| N1—C2—H2B | 109.1 | C11—C10—C3 | 118.18 (11) |
| C3—C2—H2B | 109.1 | C12—C11—C10 | 121.06 (15) |
| H2A—C2—H2B | 107.8 | C12—C11—H11 | 119.5 |
| C10—C3—C4 | 111.76 (8) | C10—C11—H11 | 119.5 |
| C10—C3—C2 | 114.52 (9) | C13—C12—C11 | 120.68 (14) |
| C4—C3—C2 | 107.62 (9) | C13—C12—H12 | 119.7 |
| C10—C3—H3 | 107.6 | C11—C12—H12 | 119.7 |
| C4—C3—H3 | 107.6 | C12—C13—C14 | 119.61 (13) |
| C2—C3—H3 | 107.6 | C12—C13—H13 | 120.2 |
| C5—C4—C9 | 118.66 (12) | C14—C13—H13 | 120.2 |
| C5—C4—C3 | 120.54 (12) | C13—C14—C15 | 120.00 (14) |
| C9—C4—C3 | 120.79 (11) | C13—C14—H14 | 120.0 |
| C4—C5—C6 | 119.52 (16) | C15—C14—H14 | 120.0 |
| C4—C5—H5 | 120.2 | C10—C15—C14 | 120.64 (12) |
| C6—C5—H5 | 120.2 | C10—C15—H15 | 119.7 |
| C7—C6—C5 | 120.67 (16) | C14—C15—H15 | 119.7 |
| C7—C6—H6 | 119.7 | ||
| C1—N1—N2—N3 | 0.47 (16) | C5—C6—C7—C8 | 0.4 (3) |
| C2—N1—N2—N3 | 178.18 (11) | C6—C7—C8—C9 | −0.7 (2) |
| N1—N2—N3—N4 | −0.49 (16) | C7—C8—C9—C4 | 0.3 (2) |
| N2—N3—N4—C1 | 0.32 (17) | C5—C4—C9—C8 | 0.42 (19) |
| N3—N4—C1—N1 | −0.01 (16) | C3—C4—C9—C8 | 179.46 (12) |
| N2—N1—C1—N4 | −0.28 (16) | C4—C3—C10—C15 | −95.17 (13) |
| C2—N1—C1—N4 | −177.76 (11) | C2—C3—C10—C15 | 27.54 (15) |
| C1—N1—C2—C3 | −118.85 (13) | C4—C3—C10—C11 | 82.93 (13) |
| N2—N1—C2—C3 | 63.98 (14) | C2—C3—C10—C11 | −154.37 (11) |
| N1—C2—C3—C10 | 60.68 (12) | C15—C10—C11—C12 | 1.0 (2) |
| N1—C2—C3—C4 | −174.40 (9) | C3—C10—C11—C12 | −177.25 (15) |
| C10—C3—C4—C5 | −116.60 (12) | C10—C11—C12—C13 | −0.2 (3) |
| C2—C3—C4—C5 | 116.84 (12) | C11—C12—C13—C14 | −0.9 (3) |
| C10—C3—C4—C9 | 64.38 (13) | C12—C13—C14—C15 | 1.3 (2) |
| C2—C3—C4—C9 | −62.18 (13) | C11—C10—C15—C14 | −0.55 (19) |
| C9—C4—C5—C6 | −0.7 (2) | C3—C10—C15—C14 | 177.55 (11) |
| C3—C4—C5—C6 | −179.78 (12) | C13—C14—C15—C10 | −0.6 (2) |
| C4—C5—C6—C7 | 0.3 (2) |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| C1—H1···N3i | 0.93 | 2.61 | 3.4690 (19) | 153 |
| C2—H2B···N4i | 0.97 | 2.50 | 3.4622 (17) | 174 |
| C2—H2A···N4ii | 0.97 | 2.56 | 3.5155 (17) | 169 |
Symmetry codes: (i) −x, y+1/2, −z+1/2; (ii) x, −y+1/2, z+1/2.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: FJ2563).
References
- Absmeier, A., Bartel, M., Carbonera, C., Jameson, G. N. L., Weinberger, P., Caneschi, A., Mereiter, K., Letard, J.-F. & Linert, W. (2006). Chem. Eur. J. 12, 2235–2243. [DOI] [PubMed]
- Bruker (2009). SAINT Bruker AXS Inc., Madison, Wisconsin, USA.
- Bruker (2011). APEX2 Bruker AXS Inc., Madison, Wisconsin, USA.
- Dowty, E. (2006). ATOMS Shape Software, Kingsport, Tennessee, USA.
- Grunert, C. M., Weinberger, P., Schweifer, J., Hampel, C., Stassen, A. F., Mereiter, K. & Linert, W. (2005). J. Mol. Struct. 733, 41–52.
- Kamiya, T. & Saito, Y. (1973). Offenlegungsschrift 2147023 (Patent).
- Macrae, C. F., Edgington, P. R., McCabe, P., Pidcock, E., Shields, G. P., Taylor, R., Towler, M. & van de Streek, J. (2006). J. Appl. Cryst. 39, 453–457.
- Momma, K. & Izumi, F. (2008). J. Appl. Cryst. 41, 653–658.
- Sheldrick, G. M. (2008a). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Sheldrick, G. M. (2008b). SADABS University of Göttingen, Germany.
- Werner, F., Mereiter, K., Tokuno, K., Inagaki, Y. & Hasegawa, M. (2009). Acta Cryst. E65, o2726–o2727. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536812027675/fj2563sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536812027675/fj2563Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536812027675/fj2563Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report







