Abstract
The title compound [systematic name: (1α,3α,6α,8β,13β,14α,16β)-20-ethyl-8-ethoxy-3,13-dihydroxy-1,6,16-trimethoxy-4-(methoxymethyl)aconitan-14-yl 4-methoxybenzoate], C35H51NO10, was isolated from roots of Aconitum carmichaeli Debx., which is a typical C19-diterpenoid alkaloid. The molecule has an aconitane carbon skeleton with four six-membered rings and two five-membered rings. The six-membered rings adopt chair conformations or boat conformations, while the five-membered rings have envelope conformations. Intramolecular O—H⋯O and O—H⋯N hydrogen bonds help to stabilize the molecular structure. Weak intermolecular C—H⋯O interactions occur in the crystal structure.
Related literature
For a related structure, see: Wang et al. (2009 ▶).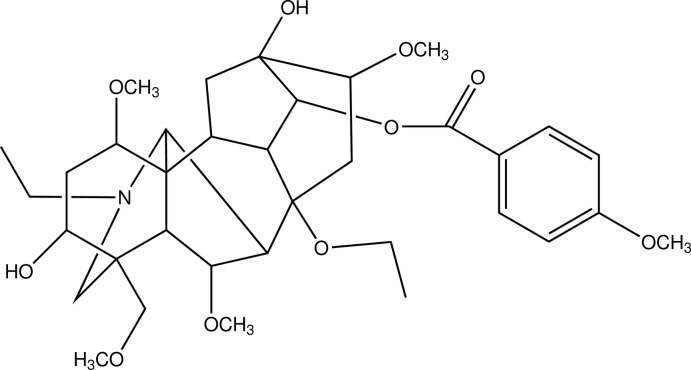
Experimental
Crystal data
C35H51NO10
M r = 645.77
Monoclinic,
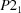
a = 10.0176 (4) Å
b = 11.7075 (5) Å
c = 14.3449 (5) Å
β = 92.528 (3)°
V = 1680.75 (11) Å3
Z = 2
Mo Kα radiation
μ = 0.09 mm−1
T = 293 K
0.41 × 0.40 × 0.38 mm
Data collection
Oxford Diffraction Xcalibur Eos diffractometer
7371 measured reflections
3609 independent reflections
2756 reflections with I > 2σ(I)
R int = 0.031
Refinement
R[F 2 > 2σ(F 2)] = 0.053
wR(F 2) = 0.131
S = 1.06
3609 reflections
438 parameters
1 restraint
H atoms treated by a mixture of independent and constrained refinement
Δρmax = 0.27 e Å−3
Δρmin = −0.17 e Å−3
Data collection: CrysAlis PRO (Oxford Diffraction, 2009 ▶); cell refinement: CrysAlis PRO; data reduction: CrysAlis PRO; program(s) used to solve structure: SHELXTL (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXTL; molecular graphics: SHELXTL; software used to prepare material for publication: SHELXTL.
Supplementary Material
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S1600536812026463/xu5540sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536812026463/xu5540Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O2—H2⋯O1 | 0.89 (6) | 2.32 (6) | 2.932 (4) | 126 (5) |
| O2—H2⋯N1 | 0.89 (6) | 2.21 (6) | 2.845 (4) | 128 (5) |
| O5—H5⋯O7 | 0.91 (5) | 1.99 (6) | 2.562 (5) | 120 (4) |
| C35—H35B⋯O2i | 0.96 | 2.56 | 3.245 (5) | 129 |
Symmetry code: (i) 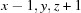 .
.
Acknowledgments
This project was supported by the Leshan Science and Technology Administration of China (12SZD128).
supplementary crystallographic information
Comment
The title compound, 8-O-ethylyunaconitine, was previously isolated from Aconitumcarmichaeli Debx., and its structure was established from the NMR and MS data. In our recent investigation, it was isolation from the root of Aconitum carmichaeli Debx, collected in the E'mei Mountain, Sichuan Province of China in August 2010, and its crystal structure was determined (Wang et al., 2009).
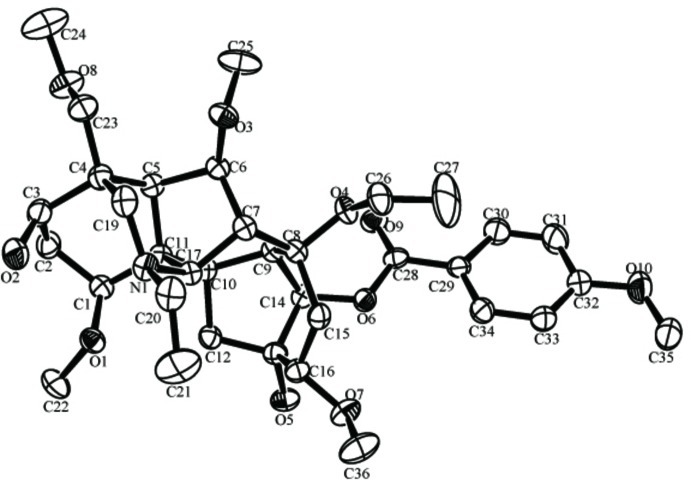
The molecular structure of the title compound is shown in Fig. 1. Six-membered rings A (C1/C2/C3/C4/C5/C11) and D (C8/C9/C14/C13/C16/C15) adopt boat conformations; six-membered ring B (C7/C8/C9/C10/C11/C17) adopts chair conformation; six-membered heterocyclic ring E (C4/C5/C11/C17/N1/C19) adopts the same chair conformation; the five-membered rings C (C9/C10/C12/C13/C14) and F (C5/C6/C7/C17/C11) display an envelope conformation, in which, the C14 and C11 act as the "envelope" respectively. The crystal structure contains intermolecular O—H···O and O—H···N hydrogen bonds. The intermolecular hydrogen bonds may be effective in the stabilization of the structure.
The absolute configuration of the title compound can not be confirmed by the present MoKa diffraction data. But it can be assumed to be the same as that reported for C19-diterpenoid alkaloids from the nature (Wang et al., 2009).
Experimental
Air-dried and powdered roots (600 g) were percolated with 0.1 M HCl (6 L). The obtained acid aqueous solution was basified with 10% aqueous NH4OH to pH 11 and then extracted with ethyl acetate (6 L × 3). Removal of the solvent under reduced pressure afforded the total crude alkaloids (5.2 g) as a yellowish amorphous powder, which was chromatographed over a silica gel column, eluting with cyclohexane-acetone (7:1→1:2) gradient system, to afford 8-O-ethylyunaconitine (256 mg). The crystals suitable for X-ray structure analysis was obtained by slow evaporation from an acetone solution at room temperature.
Refinement
moiety Hydroxyl H atoms were located in a difference Fourier map and refined isotropically. Other H atoms were located geometrically with C—H = 0.93–0.98 Å, and refined using a riding model with Uiso(H) =1.2Ueq(C). The absolute configuration has not been determined for the structure.
Figures
Fig. 1.
The molecular structure of the title compound with 30% probability displacement ellipsoids for non-H atoms.
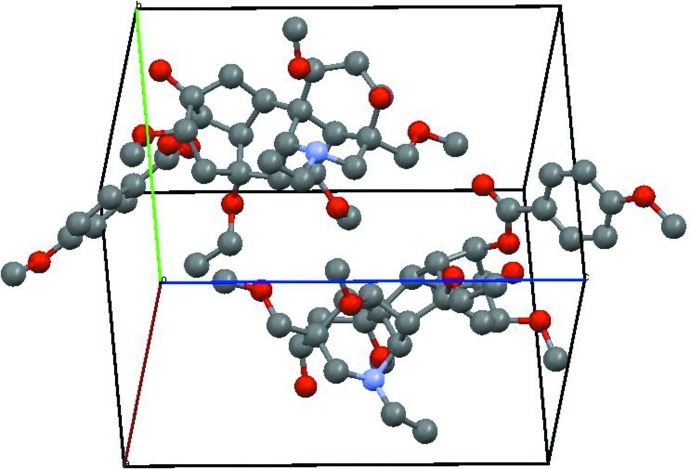
Fig. 2.
Molecular packing of the title compound.
Crystal data
| C35H51NO10 | F(000) = 696 |
| Mr = 645.77 | Dx = 1.276 Mg m−3 |
| Monoclinic, P21 | Mo Kα radiation, λ = 0.7107 Å |
| a = 10.0176 (4) Å | Cell parameters from 2465 reflections |
| b = 11.7075 (5) Å | θ = 3.1–29.1° |
| c = 14.3449 (5) Å | µ = 0.09 mm−1 |
| β = 92.528 (3)° | T = 293 K |
| V = 1680.75 (11) Å3 | Block, colorless |
| Z = 2 | 0.41 × 0.40 × 0.38 mm |
Data collection
| Oxford Diffraction Xcalibur Eos diffractometer | 2756 reflections with I > 2σ(I) |
| Radiation source: Enhance (Mo) X-ray Source | Rint = 0.031 |
| Graphite monochromator | θmax = 26.4°, θmin = 3.1° |
| Detector resolution: 16.0874 pixels mm-1 | h = −6→12 |
| ω scans | k = −13→14 |
| 7371 measured reflections | l = −17→17 |
| 3609 independent reflections |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.053 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.131 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.06 | w = 1/[σ2(Fo2) + (0.0547P)2 + 0.155P] where P = (Fo2 + 2Fc2)/3 |
| 3609 reflections | (Δ/σ)max < 0.001 |
| 438 parameters | Δρmax = 0.27 e Å−3 |
| 1 restraint | Δρmin = −0.17 e Å−3 |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O1 | 0.9158 (3) | 0.3357 (2) | 0.61833 (17) | 0.0491 (7) | |
| O2 | 0.9122 (3) | 0.2281 (3) | 0.4337 (2) | 0.0669 (9) | |
| H2 | 0.913 (6) | 0.209 (5) | 0.494 (4) | 0.10 (2)* | |
| O3 | 0.3907 (3) | 0.1776 (2) | 0.49126 (19) | 0.0521 (7) | |
| O4 | 0.3297 (3) | 0.2344 (2) | 0.73091 (18) | 0.0494 (7) | |
| O5 | 0.6836 (3) | 0.4867 (3) | 0.9172 (2) | 0.0599 (8) | |
| H5 | 0.680 (5) | 0.446 (5) | 0.971 (3) | 0.081 (18)* | |
| O6 | 0.4041 (3) | 0.4066 (3) | 0.87839 (16) | 0.0475 (7) | |
| O7 | 0.6399 (3) | 0.2797 (3) | 0.96522 (19) | 0.0672 (9) | |
| O8 | 0.5552 (4) | 0.3419 (3) | 0.3044 (2) | 0.0729 (9) | |
| O9 | 0.2353 (3) | 0.5058 (3) | 0.8094 (2) | 0.0594 (8) | |
| O10 | −0.0418 (3) | 0.2759 (4) | 1.1522 (2) | 0.0881 (12) | |
| N1 | 0.7470 (3) | 0.1413 (3) | 0.5736 (2) | 0.0465 (8) | |
| C1 | 0.8099 (4) | 0.3994 (4) | 0.5738 (2) | 0.0451 (9) | |
| H1 | 0.8112 | 0.4757 | 0.6018 | 0.054* | |
| C2 | 0.8247 (5) | 0.4144 (4) | 0.4702 (3) | 0.0565 (11) | |
| H2A | 0.7606 | 0.4707 | 0.4467 | 0.068* | |
| H2B | 0.9135 | 0.4431 | 0.4594 | 0.068* | |
| C3 | 0.8031 (4) | 0.3045 (4) | 0.4177 (3) | 0.0559 (11) | |
| H3 | 0.8009 | 0.3239 | 0.3512 | 0.067* | |
| C4 | 0.6623 (4) | 0.2487 (4) | 0.4374 (3) | 0.0474 (9) | |
| C5 | 0.5815 (4) | 0.3212 (3) | 0.5059 (2) | 0.0418 (8) | |
| H5A | 0.5515 | 0.3931 | 0.4769 | 0.050* | |
| C6 | 0.4632 (4) | 0.2573 (3) | 0.5496 (2) | 0.0411 (8) | |
| H6 | 0.3997 | 0.3151 | 0.5697 | 0.049* | |
| C7 | 0.5253 (4) | 0.1991 (3) | 0.6387 (2) | 0.0403 (8) | |
| H7 | 0.5067 | 0.1169 | 0.6376 | 0.048* | |
| C8 | 0.4703 (4) | 0.2537 (3) | 0.7257 (2) | 0.0405 (8) | |
| C9 | 0.4799 (4) | 0.3852 (3) | 0.7156 (2) | 0.0405 (9) | |
| H9 | 0.4018 | 0.4142 | 0.6795 | 0.049* | |
| C10 | 0.6107 (4) | 0.4228 (3) | 0.6692 (2) | 0.0372 (8) | |
| H10 | 0.5921 | 0.4964 | 0.6387 | 0.045* | |
| C11 | 0.6739 (3) | 0.3439 (3) | 0.5953 (2) | 0.0370 (8) | |
| C12 | 0.7097 (4) | 0.4468 (4) | 0.7546 (2) | 0.0453 (9) | |
| H12A | 0.7890 | 0.3998 | 0.7510 | 0.054* | |
| H12B | 0.7365 | 0.5264 | 0.7556 | 0.054* | |
| C13 | 0.6353 (4) | 0.4177 (4) | 0.8417 (3) | 0.0457 (9) | |
| C14 | 0.4927 (4) | 0.4450 (3) | 0.8096 (2) | 0.0425 (9) | |
| H14 | 0.4827 | 0.5276 | 0.8008 | 0.051* | |
| C15 | 0.5436 (4) | 0.2116 (3) | 0.8176 (3) | 0.0464 (9) | |
| H15A | 0.4759 | 0.1954 | 0.8621 | 0.056* | |
| H15B | 0.5866 | 0.1397 | 0.8040 | 0.056* | |
| C16 | 0.6492 (4) | 0.2893 (4) | 0.8665 (3) | 0.0493 (10) | |
| H16 | 0.7380 | 0.2633 | 0.8498 | 0.059* | |
| C17 | 0.6762 (4) | 0.2208 (3) | 0.6342 (2) | 0.0398 (8) | |
| H17 | 0.7180 | 0.2200 | 0.6972 | 0.048* | |
| C19 | 0.6822 (5) | 0.1301 (4) | 0.4802 (3) | 0.0539 (11) | |
| H19A | 0.5964 | 0.0925 | 0.4845 | 0.065* | |
| H19B | 0.7374 | 0.0841 | 0.4409 | 0.065* | |
| C20 | 0.7676 (5) | 0.0285 (4) | 0.6163 (3) | 0.0635 (13) | |
| H20A | 0.8017 | −0.0231 | 0.5701 | 0.076* | |
| H20B | 0.6821 | −0.0010 | 0.6345 | 0.076* | |
| C21 | 0.8631 (7) | 0.0297 (5) | 0.7004 (4) | 0.105 (2) | |
| H21A | 0.9388 | 0.0766 | 0.6878 | 0.157* | |
| H21B | 0.8925 | −0.0467 | 0.7141 | 0.157* | |
| H21C | 0.8187 | 0.0601 | 0.7530 | 0.157* | |
| C22 | 1.0347 (4) | 0.3991 (5) | 0.6345 (3) | 0.0698 (13) | |
| H22A | 1.0703 | 0.4202 | 0.5759 | 0.105* | |
| H22B | 1.0990 | 0.3534 | 0.6694 | 0.105* | |
| H22C | 1.0157 | 0.4668 | 0.6693 | 0.105* | |
| C23 | 0.5861 (5) | 0.2339 (4) | 0.3427 (3) | 0.0587 (11) | |
| H23A | 0.499 (5) | 0.194 (4) | 0.343 (3) | 0.063 (13)* | |
| H23B | 0.651 (4) | 0.192 (4) | 0.298 (3) | 0.054 (12)* | |
| C24 | 0.4958 (8) | 0.3360 (7) | 0.2134 (4) | 0.118 (3) | |
| H24A | 0.5539 | 0.2953 | 0.1735 | 0.177* | |
| H24B | 0.4814 | 0.4119 | 0.1897 | 0.177* | |
| H24C | 0.4118 | 0.2968 | 0.2151 | 0.177* | |
| C25 | 0.2723 (5) | 0.2212 (5) | 0.4519 (4) | 0.0827 (16) | |
| H25A | 0.2263 | 0.1625 | 0.4165 | 0.124* | |
| H25B | 0.2918 | 0.2839 | 0.4116 | 0.124* | |
| H25C | 0.2169 | 0.2474 | 0.5005 | 0.124* | |
| C26 | 0.2866 (5) | 0.1196 (4) | 0.7395 (3) | 0.0647 (13) | |
| H26A | 0.3629 | 0.0718 | 0.7564 | 0.078* | |
| H26B | 0.2493 | 0.0935 | 0.6797 | 0.078* | |
| C27 | 0.1869 (9) | 0.1074 (8) | 0.8093 (6) | 0.152 (4) | |
| H27A | 0.1193 | 0.1648 | 0.7996 | 0.228* | |
| H27B | 0.2287 | 0.1161 | 0.8704 | 0.228* | |
| H27C | 0.1468 | 0.0331 | 0.8040 | 0.228* | |
| C28 | 0.2758 (4) | 0.4412 (4) | 0.8697 (2) | 0.0443 (9) | |
| C29 | 0.1965 (4) | 0.3935 (4) | 0.9440 (2) | 0.0457 (9) | |
| C30 | 0.0640 (5) | 0.4273 (5) | 0.9512 (3) | 0.0725 (15) | |
| H30 | 0.0266 | 0.4791 | 0.9083 | 0.087* | |
| C31 | −0.0113 (5) | 0.3857 (6) | 1.0199 (4) | 0.0841 (18) | |
| H31 | −0.1000 | 0.4083 | 1.0230 | 0.101* | |
| C32 | 0.0418 (4) | 0.3107 (5) | 1.0848 (3) | 0.0608 (12) | |
| C33 | 0.1710 (4) | 0.2724 (5) | 1.0780 (3) | 0.0645 (13) | |
| H33 | 0.2063 | 0.2182 | 1.1196 | 0.077* | |
| C34 | 0.2476 (4) | 0.3163 (4) | 1.0078 (3) | 0.0577 (11) | |
| H34 | 0.3358 | 0.2926 | 1.0041 | 0.069* | |
| C35 | 0.0086 (5) | 0.1992 (5) | 1.2220 (3) | 0.0764 (15) | |
| H35A | 0.0423 | 0.1319 | 1.1929 | 0.115* | |
| H35B | −0.0618 | 0.1784 | 1.2619 | 0.115* | |
| H35C | 0.0794 | 0.2355 | 1.2582 | 0.115* | |
| C36 | 0.6919 (7) | 0.1752 (5) | 1.0015 (4) | 0.100 (2) | |
| H36A | 0.7764 | 0.1597 | 0.9751 | 0.149* | |
| H36B | 0.6306 | 0.1144 | 0.9859 | 0.149* | |
| H36C | 0.7038 | 0.1808 | 1.0681 | 0.149* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1 | 0.0442 (15) | 0.0442 (16) | 0.0588 (15) | −0.0045 (13) | 0.0015 (12) | 0.0027 (14) |
| O2 | 0.0565 (19) | 0.077 (2) | 0.068 (2) | 0.0097 (19) | 0.0158 (15) | −0.0040 (19) |
| O3 | 0.0478 (16) | 0.0418 (16) | 0.0661 (16) | −0.0040 (13) | −0.0061 (13) | −0.0134 (14) |
| O4 | 0.0397 (14) | 0.0444 (15) | 0.0647 (16) | −0.0053 (13) | 0.0112 (12) | 0.0001 (14) |
| O5 | 0.068 (2) | 0.060 (2) | 0.0502 (16) | −0.0028 (17) | −0.0112 (14) | −0.0109 (16) |
| O6 | 0.0440 (14) | 0.0554 (17) | 0.0433 (13) | 0.0083 (14) | 0.0046 (11) | 0.0043 (13) |
| O7 | 0.086 (2) | 0.067 (2) | 0.0473 (15) | 0.0082 (19) | −0.0039 (15) | 0.0120 (15) |
| O8 | 0.099 (3) | 0.065 (2) | 0.0528 (16) | 0.001 (2) | −0.0128 (16) | −0.0016 (17) |
| O9 | 0.0577 (18) | 0.0659 (19) | 0.0540 (16) | 0.0144 (16) | −0.0042 (13) | 0.0077 (15) |
| O10 | 0.063 (2) | 0.122 (4) | 0.081 (2) | 0.006 (2) | 0.0232 (17) | 0.026 (2) |
| N1 | 0.054 (2) | 0.0347 (18) | 0.0518 (18) | 0.0082 (16) | 0.0081 (15) | −0.0010 (15) |
| C1 | 0.045 (2) | 0.040 (2) | 0.050 (2) | −0.0021 (18) | 0.0019 (16) | −0.0039 (18) |
| C2 | 0.058 (3) | 0.056 (3) | 0.056 (2) | −0.004 (2) | 0.0084 (19) | 0.009 (2) |
| C3 | 0.058 (3) | 0.063 (3) | 0.048 (2) | 0.003 (2) | 0.0096 (18) | 0.002 (2) |
| C4 | 0.051 (2) | 0.046 (2) | 0.0460 (19) | 0.000 (2) | 0.0048 (16) | −0.0061 (18) |
| C5 | 0.049 (2) | 0.0318 (19) | 0.0441 (18) | −0.0024 (18) | 0.0002 (16) | 0.0022 (17) |
| C6 | 0.0419 (19) | 0.0340 (19) | 0.0474 (19) | −0.0031 (17) | 0.0014 (15) | −0.0055 (17) |
| C7 | 0.043 (2) | 0.0269 (18) | 0.051 (2) | −0.0019 (16) | 0.0044 (16) | −0.0024 (16) |
| C8 | 0.0381 (19) | 0.033 (2) | 0.050 (2) | −0.0009 (16) | 0.0018 (15) | 0.0008 (17) |
| C9 | 0.041 (2) | 0.036 (2) | 0.0446 (19) | 0.0041 (17) | 0.0035 (15) | −0.0027 (16) |
| C10 | 0.0421 (19) | 0.0269 (18) | 0.0429 (17) | −0.0009 (16) | 0.0065 (15) | 0.0006 (15) |
| C11 | 0.0391 (19) | 0.0306 (18) | 0.0410 (17) | −0.0024 (16) | −0.0007 (14) | −0.0006 (15) |
| C12 | 0.046 (2) | 0.041 (2) | 0.050 (2) | −0.0084 (18) | 0.0030 (16) | −0.0012 (18) |
| C13 | 0.046 (2) | 0.046 (2) | 0.0450 (19) | 0.0033 (19) | −0.0018 (16) | −0.0037 (18) |
| C14 | 0.050 (2) | 0.0351 (19) | 0.0427 (19) | 0.0018 (18) | 0.0045 (16) | −0.0020 (17) |
| C15 | 0.049 (2) | 0.041 (2) | 0.050 (2) | 0.0055 (19) | 0.0087 (17) | 0.0095 (18) |
| C16 | 0.049 (2) | 0.051 (3) | 0.048 (2) | 0.005 (2) | −0.0001 (17) | 0.0098 (19) |
| C17 | 0.043 (2) | 0.0332 (19) | 0.0431 (18) | 0.0003 (17) | 0.0022 (15) | −0.0013 (16) |
| C19 | 0.060 (3) | 0.040 (2) | 0.063 (2) | 0.003 (2) | 0.016 (2) | −0.007 (2) |
| C20 | 0.074 (3) | 0.041 (2) | 0.077 (3) | 0.018 (2) | 0.018 (2) | 0.003 (2) |
| C21 | 0.139 (6) | 0.073 (4) | 0.100 (4) | 0.046 (4) | −0.024 (4) | 0.020 (3) |
| C22 | 0.046 (2) | 0.073 (3) | 0.091 (3) | −0.017 (3) | 0.003 (2) | −0.006 (3) |
| C23 | 0.070 (3) | 0.053 (3) | 0.052 (2) | −0.002 (3) | −0.001 (2) | −0.014 (2) |
| C24 | 0.181 (8) | 0.097 (5) | 0.071 (3) | −0.002 (6) | −0.042 (4) | −0.008 (4) |
| C25 | 0.081 (3) | 0.051 (3) | 0.112 (4) | 0.000 (3) | −0.047 (3) | −0.021 (3) |
| C26 | 0.064 (3) | 0.053 (3) | 0.079 (3) | −0.019 (2) | 0.016 (2) | −0.002 (2) |
| C27 | 0.167 (8) | 0.120 (6) | 0.175 (7) | −0.090 (6) | 0.083 (6) | −0.012 (6) |
| C28 | 0.051 (2) | 0.044 (2) | 0.0372 (18) | 0.0057 (19) | −0.0053 (16) | −0.0086 (18) |
| C29 | 0.043 (2) | 0.053 (2) | 0.0401 (17) | 0.0025 (19) | −0.0028 (15) | −0.0024 (18) |
| C30 | 0.052 (3) | 0.094 (4) | 0.072 (3) | 0.018 (3) | 0.004 (2) | 0.023 (3) |
| C31 | 0.049 (3) | 0.112 (5) | 0.092 (4) | 0.021 (3) | 0.014 (2) | 0.029 (4) |
| C32 | 0.053 (2) | 0.074 (3) | 0.055 (2) | −0.005 (3) | 0.0042 (19) | 0.005 (2) |
| C33 | 0.053 (3) | 0.083 (4) | 0.058 (2) | 0.007 (3) | 0.001 (2) | 0.015 (3) |
| C34 | 0.045 (2) | 0.073 (3) | 0.056 (2) | 0.012 (2) | 0.0083 (18) | 0.005 (2) |
| C35 | 0.084 (4) | 0.080 (4) | 0.066 (3) | −0.009 (3) | 0.016 (3) | 0.005 (3) |
| C36 | 0.139 (6) | 0.083 (4) | 0.074 (3) | 0.019 (4) | −0.023 (3) | 0.036 (3) |
Geometric parameters (Å, º)
| O1—C1 | 1.425 (5) | C12—C13 | 1.521 (5) |
| O1—C22 | 1.414 (5) | C13—C14 | 1.516 (5) |
| O2—H2 | 0.89 (6) | C13—C16 | 1.550 (6) |
| O2—C3 | 1.424 (5) | C14—H14 | 0.9800 |
| O3—C6 | 1.430 (4) | C15—H15A | 0.9700 |
| O3—C25 | 1.388 (6) | C15—H15B | 0.9700 |
| O4—C8 | 1.431 (4) | C15—C16 | 1.541 (6) |
| O4—C26 | 1.419 (5) | C16—H16 | 0.9800 |
| O5—H5 | 0.91 (5) | C17—H17 | 0.9800 |
| O5—C13 | 1.419 (5) | C19—H19A | 0.9700 |
| O6—C14 | 1.429 (4) | C19—H19B | 0.9700 |
| O6—C28 | 1.348 (5) | C20—H20A | 0.9700 |
| O7—C16 | 1.427 (4) | C20—H20B | 0.9700 |
| O7—C36 | 1.419 (6) | C20—C21 | 1.507 (7) |
| O8—C23 | 1.407 (6) | C21—H21A | 0.9600 |
| O8—C24 | 1.412 (6) | C21—H21B | 0.9600 |
| O9—C28 | 1.206 (5) | C21—H21C | 0.9600 |
| O10—C32 | 1.369 (5) | C22—H22A | 0.9600 |
| O10—C35 | 1.420 (6) | C22—H22B | 0.9600 |
| N1—C17 | 1.475 (5) | C22—H22C | 0.9600 |
| N1—C19 | 1.469 (5) | C23—H23A | 0.99 (5) |
| N1—C20 | 1.466 (5) | C23—H23B | 1.05 (4) |
| C1—H1 | 0.9800 | C24—H24A | 0.9600 |
| C1—C2 | 1.511 (5) | C24—H24B | 0.9600 |
| C1—C11 | 1.553 (5) | C24—H24C | 0.9600 |
| C2—H2A | 0.9700 | C25—H25A | 0.9600 |
| C2—H2B | 0.9700 | C25—H25B | 0.9600 |
| C2—C3 | 1.502 (6) | C25—H25C | 0.9600 |
| C3—H3 | 0.9800 | C26—H26A | 0.9700 |
| C3—C4 | 1.590 (6) | C26—H26B | 0.9700 |
| C4—C5 | 1.553 (5) | C26—C27 | 1.452 (8) |
| C4—C19 | 1.528 (6) | C27—H27A | 0.9600 |
| C4—C23 | 1.539 (6) | C27—H27B | 0.9600 |
| C5—H5A | 0.9800 | C27—H27C | 0.9600 |
| C5—C6 | 1.556 (5) | C28—C29 | 1.468 (5) |
| C5—C11 | 1.571 (5) | C29—C30 | 1.394 (6) |
| C6—H6 | 0.9800 | C29—C34 | 1.369 (6) |
| C6—C7 | 1.554 (5) | C30—H30 | 0.9300 |
| C7—H7 | 0.9800 | C30—C31 | 1.359 (7) |
| C7—C8 | 1.528 (5) | C31—H31 | 0.9300 |
| C7—C17 | 1.537 (5) | C31—C32 | 1.370 (7) |
| C8—C9 | 1.549 (5) | C32—C33 | 1.377 (6) |
| C8—C15 | 1.560 (5) | C33—H33 | 0.9300 |
| C9—H9 | 0.9800 | C33—C34 | 1.391 (6) |
| C9—C10 | 1.559 (5) | C34—H34 | 0.9300 |
| C9—C14 | 1.520 (5) | C35—H35A | 0.9600 |
| C10—H10 | 0.9800 | C35—H35B | 0.9600 |
| C10—C11 | 1.561 (5) | C35—H35C | 0.9600 |
| C10—C12 | 1.568 (5) | C36—H36A | 0.9600 |
| C11—C17 | 1.545 (5) | C36—H36B | 0.9600 |
| C12—H12A | 0.9700 | C36—H36C | 0.9600 |
| C12—H12B | 0.9700 | ||
| C22—O1—C1 | 113.9 (3) | C16—C15—H15B | 107.5 |
| C3—O2—H2 | 107 (4) | O7—C16—C13 | 107.2 (3) |
| C25—O3—C6 | 113.6 (3) | O7—C16—C15 | 109.4 (3) |
| C26—O4—C8 | 117.3 (3) | O7—C16—H16 | 108.6 |
| C13—O5—H5 | 109 (3) | C13—C16—H16 | 108.6 |
| C28—O6—C14 | 117.3 (3) | C15—C16—C13 | 114.5 (3) |
| C36—O7—C16 | 113.1 (4) | C15—C16—H16 | 108.6 |
| C23—O8—C24 | 113.2 (4) | N1—C17—C7 | 114.8 (3) |
| C32—O10—C35 | 118.4 (4) | N1—C17—C11 | 112.1 (3) |
| C19—N1—C17 | 112.9 (3) | N1—C17—H17 | 109.9 |
| C20—N1—C17 | 112.7 (3) | C7—C17—C11 | 99.8 (3) |
| C20—N1—C19 | 110.4 (3) | C7—C17—H17 | 109.9 |
| O1—C1—H1 | 107.3 | C11—C17—H17 | 109.9 |
| O1—C1—C2 | 113.5 (3) | N1—C19—C4 | 109.3 (3) |
| O1—C1—C11 | 109.6 (3) | N1—C19—H19A | 109.8 |
| C2—C1—H1 | 107.3 | N1—C19—H19B | 109.8 |
| C2—C1—C11 | 111.7 (3) | C4—C19—H19A | 109.8 |
| C11—C1—H1 | 107.3 | C4—C19—H19B | 109.8 |
| C1—C2—H2A | 109.2 | H19A—C19—H19B | 108.3 |
| C1—C2—H2B | 109.2 | N1—C20—H20A | 108.9 |
| H2A—C2—H2B | 107.9 | N1—C20—H20B | 108.9 |
| C3—C2—C1 | 112.1 (4) | N1—C20—C21 | 113.5 (4) |
| C3—C2—H2A | 109.2 | H20A—C20—H20B | 107.7 |
| C3—C2—H2B | 109.2 | C21—C20—H20A | 108.9 |
| O2—C3—C2 | 111.4 (4) | C21—C20—H20B | 108.9 |
| O2—C3—H3 | 106.7 | C20—C21—H21A | 109.5 |
| O2—C3—C4 | 113.2 (4) | C20—C21—H21B | 109.5 |
| C2—C3—H3 | 106.7 | C20—C21—H21C | 109.5 |
| C2—C3—C4 | 111.8 (3) | H21A—C21—H21B | 109.5 |
| C4—C3—H3 | 106.7 | H21A—C21—H21C | 109.5 |
| C5—C4—C3 | 112.3 (3) | H21B—C21—H21C | 109.5 |
| C19—C4—C3 | 110.1 (4) | O1—C22—H22A | 109.5 |
| C19—C4—C5 | 107.7 (3) | O1—C22—H22B | 109.5 |
| C19—C4—C23 | 107.6 (4) | O1—C22—H22C | 109.5 |
| C23—C4—C3 | 107.4 (3) | H22A—C22—H22B | 109.5 |
| C23—C4—C5 | 111.6 (3) | H22A—C22—H22C | 109.5 |
| C4—C5—H5A | 111.1 | H22B—C22—H22C | 109.5 |
| C4—C5—C6 | 114.9 (3) | O8—C23—C4 | 109.6 (4) |
| C4—C5—C11 | 107.7 (3) | O8—C23—H23A | 104 (3) |
| C6—C5—H5A | 111.1 | O8—C23—H23B | 108 (2) |
| C6—C5—C11 | 100.6 (3) | C4—C23—H23A | 117 (2) |
| C11—C5—H5A | 111.1 | C4—C23—H23B | 107 (2) |
| O3—C6—C5 | 117.0 (3) | H23A—C23—H23B | 111 (4) |
| O3—C6—H6 | 107.6 | O8—C24—H24A | 109.5 |
| O3—C6—C7 | 111.7 (3) | O8—C24—H24B | 109.5 |
| C5—C6—H6 | 107.6 | O8—C24—H24C | 109.5 |
| C7—C6—C5 | 104.9 (3) | H24A—C24—H24B | 109.5 |
| C7—C6—H6 | 107.6 | H24A—C24—H24C | 109.5 |
| C6—C7—H7 | 110.4 | H24B—C24—H24C | 109.5 |
| C8—C7—C6 | 110.1 (3) | O3—C25—H25A | 109.5 |
| C8—C7—H7 | 110.4 | O3—C25—H25B | 109.5 |
| C8—C7—C17 | 110.9 (3) | O3—C25—H25C | 109.5 |
| C17—C7—C6 | 104.6 (3) | H25A—C25—H25B | 109.5 |
| C17—C7—H7 | 110.4 | H25A—C25—H25C | 109.5 |
| O4—C8—C7 | 111.6 (3) | H25B—C25—H25C | 109.5 |
| O4—C8—C9 | 103.1 (3) | O4—C26—H26A | 109.2 |
| O4—C8—C15 | 109.5 (3) | O4—C26—H26B | 109.2 |
| C7—C8—C9 | 108.2 (3) | O4—C26—C27 | 112.0 (5) |
| C7—C8—C15 | 112.7 (3) | H26A—C26—H26B | 107.9 |
| C9—C8—C15 | 111.4 (3) | C27—C26—H26A | 109.2 |
| C8—C9—H9 | 110.0 | C27—C26—H26B | 109.2 |
| C8—C9—C10 | 112.2 (3) | C26—C27—H27A | 109.5 |
| C10—C9—H9 | 110.0 | C26—C27—H27B | 109.5 |
| C14—C9—C8 | 112.2 (3) | C26—C27—H27C | 109.5 |
| C14—C9—H9 | 110.0 | H27A—C27—H27B | 109.5 |
| C14—C9—C10 | 102.1 (3) | H27A—C27—H27C | 109.5 |
| C9—C10—H10 | 107.0 | H27B—C27—H27C | 109.5 |
| C9—C10—C11 | 119.7 (3) | O6—C28—C29 | 111.2 (3) |
| C9—C10—C12 | 103.4 (3) | O9—C28—O6 | 123.0 (4) |
| C11—C10—H10 | 107.0 | O9—C28—C29 | 125.7 (4) |
| C11—C10—C12 | 112.0 (3) | C30—C29—C28 | 119.5 (4) |
| C12—C10—H10 | 107.0 | C34—C29—C28 | 122.6 (4) |
| C1—C11—C5 | 113.5 (3) | C34—C29—C30 | 117.9 (4) |
| C1—C11—C10 | 105.8 (3) | C29—C30—H30 | 119.6 |
| C10—C11—C5 | 114.4 (3) | C31—C30—C29 | 120.8 (5) |
| C17—C11—C1 | 117.6 (3) | C31—C30—H30 | 119.6 |
| C17—C11—C5 | 97.8 (3) | C30—C31—H31 | 119.6 |
| C17—C11—C10 | 107.9 (3) | C30—C31—C32 | 120.8 (5) |
| C10—C12—H12A | 110.4 | C32—C31—H31 | 119.6 |
| C10—C12—H12B | 110.4 | O10—C32—C31 | 116.0 (4) |
| H12A—C12—H12B | 108.6 | O10—C32—C33 | 124.0 (4) |
| C13—C12—C10 | 106.6 (3) | C31—C32—C33 | 120.0 (4) |
| C13—C12—H12A | 110.4 | C32—C33—H33 | 120.6 |
| C13—C12—H12B | 110.4 | C32—C33—C34 | 118.7 (4) |
| O5—C13—C12 | 109.6 (3) | C34—C33—H33 | 120.6 |
| O5—C13—C14 | 113.4 (3) | C29—C34—C33 | 121.8 (4) |
| O5—C13—C16 | 110.6 (3) | C29—C34—H34 | 119.1 |
| C12—C13—C16 | 111.3 (3) | C33—C34—H34 | 119.1 |
| C14—C13—C12 | 101.1 (3) | O10—C35—H35A | 109.5 |
| C14—C13—C16 | 110.4 (3) | O10—C35—H35B | 109.5 |
| O6—C14—C9 | 115.9 (3) | O10—C35—H35C | 109.5 |
| O6—C14—C13 | 109.2 (3) | H35A—C35—H35B | 109.5 |
| O6—C14—H14 | 109.7 | H35A—C35—H35C | 109.5 |
| C9—C14—H14 | 109.7 | H35B—C35—H35C | 109.5 |
| C13—C14—C9 | 102.3 (3) | O7—C36—H36A | 109.5 |
| C13—C14—H14 | 109.7 | O7—C36—H36B | 109.5 |
| C8—C15—H15A | 107.5 | O7—C36—H36C | 109.5 |
| C8—C15—H15B | 107.5 | H36A—C36—H36B | 109.5 |
| H15A—C15—H15B | 107.0 | H36A—C36—H36C | 109.5 |
| C16—C15—C8 | 119.1 (3) | H36B—C36—H36C | 109.5 |
| C16—C15—H15A | 107.5 |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O2—H2···O1 | 0.89 (6) | 2.32 (6) | 2.932 (4) | 126 (5) |
| O2—H2···N1 | 0.89 (6) | 2.21 (6) | 2.845 (4) | 128 (5) |
| O5—H5···O7 | 0.91 (5) | 1.99 (6) | 2.562 (5) | 120 (4) |
| C35—H35B···O2i | 0.96 | 2.56 | 3.245 (5) | 129 |
Symmetry code: (i) x−1, y, z+1.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: XU5540).
References
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S1600536812026463/xu5540sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536812026463/xu5540Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report