Abstract
Regulators of G-protein signaling (RGS proteins) comprise a large family of signal transduction molecules that modulate G-protein-coupled-receptor (GPCR) function. Among the RGS proteins expressed in the brain, RGS9-2 is very abundant in the striatum, a brain region involved in movement, motivation, mood and addiction. This protein negatively modulates signal transduction thus playing a key part in striatal function and resultant behavioral responses. In particular, there is evidence of important interactions with μ-opioid- and dopamine D2-receptor signaling pathways. Several studies indicate that manipulations of RGS9-2 levels in the striatum might greatly affect pharmacological responses. These findings indicate that treatment strategies targeting RGS9-2 levels or activity might be used to enhance responses to drugs acting at GPCRs and/or prevent undesired drug actions.
G-protein-coupled receptors in the striatum as therapeutic targets
The striatal complex, which consists of medium spiny neurons and cholinergic interneurons, mediates important physiological functions including mood, motivation, reward, motor activity and some aspects of learning and memory [1]. Dysfunctions in striatal neurons are linked to diseases such as Parkinson’s, Huntington’s, depression and addiction [2–4]. G-protein-coupled receptors (GPCRs) have an essential role in neuronal responses in the striatum and comprise important pharmacological targets for several central nervous system disorders [5]. To date, few of the currently available medications target particular receptor types or neural networks, and this lack of selectivity often leads to the development of serious side effects. Several new studies provide evidence that interventions in signal transduction events that follow activation of a receptor can improve the therapeutic effects of a drug or prevent some undesired actions.
The diverse family of RGS proteins
The discovery of RGS proteins (regulators of G-protein signaling) has provided a better understanding of the mechanisms controlling GPCR responsiveness and deactivation. Sst2 was the first RGS-like protein discovered in yeast as a negative regulator acting at the level of Gα subunits [6]. Since then, a large number of studies have shown that RGS proteins bind to activated Gα subunits via a conserved 125 amino acid domain and accelerate GTP hydrolysis [7]. This action accelerates signaling termination by promoting the return of the Gα subunit to the GDP-inactive state. In several cases, RGS proteins might negatively modulate signaling by competing with Gα subunits for binding to effector molecules (such as phospholipase C β) independently of their GTPase action [8]. Members of the RGS family also seem to control intracellular calcium levels via direct interactions with ion channels [9]. Several studies indicate regional and functional specificity for most members of the RGS family. This specificity derives from the structural diversity among RGSs (Box 1), the different tissue distribution patterns (Table 1) and selectivity for particular Gα subunits [10,11]. It is also becoming evident that RGS proteins are part of macromolecular complexes formed upon activation of particular receptors [12,13].
Box 1. The RGS family of proteins.
Regulators of G-protein signaling (RGS) proteins comprise a family of intracellular proteins that negatively modulate signaling through G-protein-coupled receptors (GPCRs). RGS proteins are defined by the presence of a conserved 120 amino acid RGS consensus domain. This domain binds the activated GTP-bound Gα subunit of heterotrimeric G proteins and dramatically accelerates the rate of GTP hydrolysis. Thus, by acting as GTPase-accelerating proteins (GAPs), RGS proteins speed the return to the inactive GDP-bound Gα with subsequent recoupling to, and inactivation of, Gβγ subunits, thus terminating downstream signaling. Because G-protein activation is a balance between the rate of binding of GTP, which is stimulated by agonist-occupied receptor, and the rate of hydrolysis of GTP, which is accelerated by the GAP activity, the RGS proteins are intimately involved in determining the efficiency of receptor-stimulated G-protein signaling.
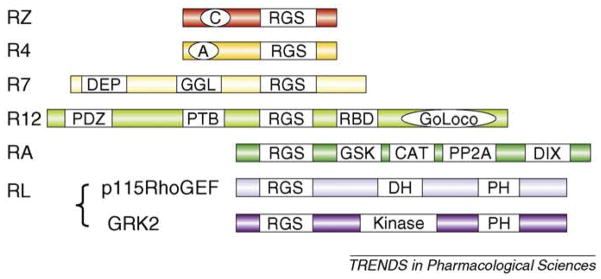
There have been 37 proteins identified with the RGS consensus domains. These bind Gα and most have efficient GAP activity and have been shown to inhibit G-protein signaling. In addition to the Gα subunit specificity, there is evidence for receptor specificity of RGS protein function, although an important determinant of selectivity might be their distribution. The RGS family has been subdivided on the basis of amino acid composition of the RGS domain. The RZ and R4 subgroups of proteins consist primarily of the RGS domain with short N and C termini. Other families are much larger structures with subfamily-specific domains for protein–protein interactions that function in scaffolding, targeting and regulation (Figure I). In addition to the ‘classical’ RGS proteins, proteins containing weak RGS homology (RH) domains that interact with Gα proteins have been identified and comprise a rather miscellaneous (RL) group. For example, in addition to interacting with GPCRs via its kinase domain, the RH domain in GPCR kinase 2 (GRK2) binds to Gαq and inhibits its ability to activate phospholipase C. By contrast, the Rho guanine nucleotide exchange factors (RhoGEFs) members of the RGS family bind somewhat selectively to Gα12 and Gα13 and serve not as negative modulators but rather as effectors transferring information from the GPCR and activated Gα subunits to activation of Rho signaling. RGS proteins can be post-translationally modified by phosphorylation and/or palmitoylatin, which can alter GAP activity, cellular localization and interaction with other proteins.
Figure I.
RGS protein subfamilies. The RZ family includes RGS17 (also known as RGSZ2), RGS20 (RGSZ1) and RGS19 (GAIP). The R4 family comprises a large number of proteins (i.e. RGS1, RGS3, RGS2, RGS4, RGS8, RGS13, RGS16 and RGS18). The RZ and R4 families are GAPs with either a poly-cysteine region (C) that is likely to be reversibly palmitoylated or an amphipathic helix (A). The N-terminal region of the R4 family seems to be important for receptor selectivity. The R7 family comprising RGS6, RGS7, RGS9 and RGS11 is more complex with protein-binding domains. The N-terminal DEP binds to R7BP and the GGL (Gγ-like) domain binds to Gβ5. The R12 family comprises RGS10, RGS12 and RGS14. RGS12 and RGS14 are larger with a RBD (Rap1/2-binding) domain and a GoLoco motif that can bind to Gα to displace Gβγ and inhibit guanine nucleotide release; RGS12 has additional N-terminal protein-binding domains (PTBs). Members of the RA family (axin and conductin) act as scaffolds binding the transcription factor β-cartenin (CAT) and controlling its phosphorylation through binding domains for glycogen synthase kinase (GSK3β) and protein phosphatase 2A (PP2A); there is also a dimerization domain (DIX). The RL subfamily is diverse and the RGS domain has low homology. Important examples include GRK2 and p115RhoGEF with a PH (pleckstrin homology) domain and either a DH (dbl homology) or kinase domain, respectively.
Table 1.
Regional mRNA expression of RGS proteins in mouse braina
| Brain region | mRNA expression |
|---|---|
| Cortex | RGS4 > RGS10 > RGS8 > RGS7 > RGS6 |
| Nucleus accumbens | RGS9-2 ≫ RGS8 > RGS4 > RGS7 |
| Dorsal striatum | RGS9-2 ≫ RGS8 > RGS4 > RGS7, RGS10, RGS5 |
| Hippocampus | RGS10 > RGS7 > RGS4, RGS5, RGS8 |
| Amygdala | RGS5 > RGS4 > RGS7 |
| Thalamus | RGS4 > RGS8 > RGS7 > RGS5, RGS6 |
| Hypothalamus | RGS5 > RGS4 > RGS7 > RGS10 > RGS9 |
| Ventral tegmental area | RGS8 > RGS6, RGS7 |
| Dorsal raphe | RGS10 > RGS8 > RGS7 |
| Locus coeruleus | RGS4, RGS7 > RGS8, RGS3, RGS6 |
| Cerebellum | RGS8, RGS7 |
Table created from data taken from Ref. [10].
A fundamental role of RGS proteins in physiological functions has been identified in yeast, Caenorhabditis elegans, Drosophila and mammals. Initial studies in C. elegans demonstrated that RGS proteins modulate locomotor activity via actions at D1-like and D2-like dopamine receptors [14]. In rodents, RGS proteins were shown to have an important role in cardiovascular function, learning and memory, stress responses, analgesia and addiction [11,15]. Genetic studies in humans have linked RGS4 to schizophrenia [16] and RGS2 to hypertension and anxiety [15]. Postmortem studies identified increased RGS9-2 levels in brain tissue from Parkinson’s patients [15]. Finally, loss of RGS9 function in the retina was shown to lead to bradyopsia, a condition characterized by inability to see moving objects during sudden changes in light intensity [17].
RGS9-2 structure and distribution
Members of the R7 subfamily (RGS9-1, RGS9-2, RGS7, RGS6 and RGS11) are among the larger RGS proteins and share common structural features. In addition to the conserved RGS domain, common features include a DEP (disheveled, EGL-10, pleckstrin) domain, which has a role in membrane association [18] and binding to adaptor proteins such as R9AP (RGS9-1 anchor protein) in the retina or R7BP (R7 binding protein) in the brain [15]. The R7 family members also share a GGL (Gγ-like) domain that enables binding to the G-protein β5 subunit (Gβ5) [19]. Binding to R7BP (or R9AP in the retina) and Gβ5 is essential for protein stability and localization [20,21]. Members of the R7 family differ in both GTPase activity and brain expression patterns [10,21] which might enable receptor-specific regulation in the central nervous system.
RGS9-2, the brain-specific splice variant of the RGS9 gene [22] (Figure 1), is very abundant in the striatum over other brain regions and modulates responses to several GPCRs [10,22–25] (Figure 2).
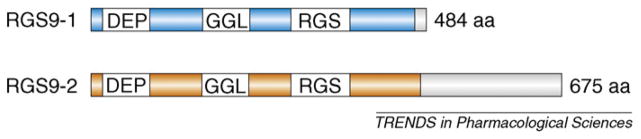
Figure 1.
Structural organization of the RGS9-1 and RGS9-2 proteins. Both RGS9-1 and RGS9-2 proteins contain an N-terminal disheveled, EGL10, pleckstrin homology (DEP) domain, a G-protein γ-subunit-like (GGL) domain and the regulator of G-protein signaling (RGS) domain. RGS9-2 contains an additional C-terminal phosphodiesterase-γ-like domain. RGS9-1 is abundant in the retina, whereas RGS9-2 is highly expressed in the striatum. Abbreviations: aa, amino acids.
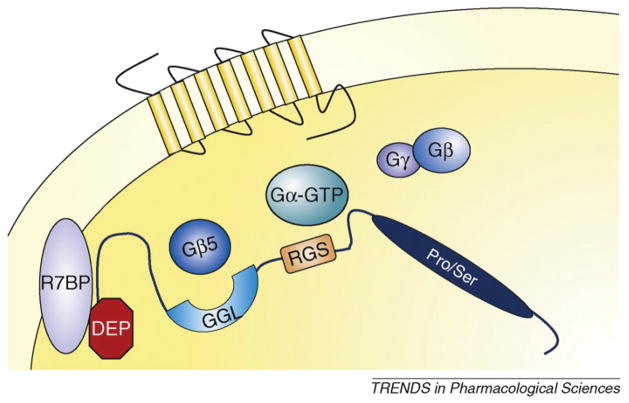
Figure 2.
RGS9-2 complexes modulate GPCR signaling. Interactions with Gβ5 and with R7BP (R7 binding protein) are essential for membrane localization and stability of RGS9-2. Via the RGS domain, RGS9-2 associates with activated Gα subunits and accelerates GTP hydrolysis. This action promotes reassociation of the Gα subunit with the βγ complex and signaling termination.
RGS9-2 is a potent modulator of psychostimulant and opioid drug actions
Behavioral phenotyping of RGS9-gene knockout (RGS9KO) mice revealed that these mice perform similarly to their wild-type controls (RGS9WT) in tests assessing cognitive function, auditory gating and general behavioral reactivity. Specifically, RGS9KO mice showed normal cue and contextual fear conditioning, pre-pulse inhibition and acoustic-startle thresholds [26]. Interestingly, although adult RGS9KO mice showed no motor deficits, older animals (12–14-month-old males) exhibited compromised motor coordination in the rotarod paradigm [26]. The cellular basis for the aging deficit has not been investigated, but it is thought to be related to compromised cholinergic and dopaminergic signaling in the striatum.
The unique localization of RGS9-2, especially in the nucleus accumbens (NAc) of the dopamine reward pathway, gave the first clue that this RGS protein might be involved in addiction. The first real evidence came from studies showing altered regulation of RGS9-2 protein levels after acute or repeated exposure to drugs of abuse. RGS9-2 levels in the striatum of rats self-administering cocaine were decreased compared with those of non-addicted controls [23]. Behavioral analysis of RGS9KO mice provided further evidence for the role of RGS9-2 in drug addiction. The locomotor-activating actions of dopaminergic or opioidergic agonists such as cocaine, amphetamine and morphine were more pronounced in RGS9KO mice [23,25] compared with RGS9WT. The role of RGS9-2 in the rewarding actions of psychostimulants has been shown by studies using place conditioning and locomotor sensitization assays [23]. Lack of the RGS9 gene led to accelerated locomotor sensitization and increased reward sensitivity.
Similar observations were made in studies examining the role of RGS9-2 in opiate addiction. Indeed, RGS9KO mice showed morphine reward at drug doses 10 times lower than those required for their wild-type littermate controls [25]. This increased sensitivity to the rewarding actions of morphine is attributed to RGS9-2 actions in the NAc. Restoring levels of RGS9-2 in the NAc using viral-mediated gene transfer brings reward thresholds close to levels of wild-type animals. In addition, RGS9KO mice show increased sensitivity to morphine in behavioral tests assessing spinal and supraspinal analgesia. The increased opioid analgesic sensitivity resulted from modulation of μ-opioid-receptor signaling by RGS9-2-containing complexes [27]. The studies on the RGS9KO mice also demonstrated that RGS9-2 potently affected other aspects of opiate addiction such as dependence and analgesic tolerance. Ongoing studies are investigating whether knockdown of RGS9-2 activity improves the actions of opiate analgesics under chronic pain conditions. Using a sciatic-nerve-injury model in mice, we demonstrate that RGS9KO mice respond better to the analgesic effects of morphine and develop morphine tolerance much later than wild-type controls* (Table 2).
Table 2.
Basal and induced behavioral analysis of RGS9-manipulated animalsa
| Behavior | Animal model | Manipulation | Treatment | Behavioral test | Behavioral phenotype relative to control | Refs |
|---|---|---|---|---|---|---|
| Coordination or cognition | ||||||
|
|
||||||
| Gross motor coordination | RGS9KO mice | None | None | Accelerating rotarod | Deficits in motor coordination; similar rate of acute motor learning | [26] |
|
|
||||||
| None | Locomotor activity | Similar horizontal locomotor activity | [26] | |||
|
|
||||||
| Emotional behaviors | RGS9KO mice | None | None | Fear conditioning | Similar context and cue-dependent fear conditioning | [26] |
|
|
||||||
| None | Open field | Similar anxiety-like behavior | [26] | |||
|
|
||||||
| None | Pre-pulse inhibition | Similar sensory motor gating | [26] | |||
|
|
||||||
| None | Shock threshold | Similar startle amplitude | [26] | |||
|
| ||||||
| Motor behavior | ||||||
| Locomotor response to dopamine-receptor agonists | Rats | Unilateral HSV–RGS9-2 (versus HSV–LacZ) overexpression in the NAc | Acute apomorphine (dopamine-receptor agonist) | Circling and rotation recording | ↑ ↑b Rotational bias towards the side of overexpression | [23] |
|
|
||||||
| Acute quinpirole (D2-receptor agonist) | Circling and rotation recording | ↑ Rotational bias towards the side of overexpression | [23] | |||
|
|
||||||
| Acute SKF81297 (D1-receptor agonist) | Circling and rotation recording | No rotational bias | [23] | |||
|
|
||||||
| Antipsychotic drug-induced dyskinesia | RGS9KO mice | None | Acute apomorphine, or quinpirole or SKF38393 | AIM recording | Similar induction of few AIMs | [24] |
|
|
||||||
| 3 days reserpine | AIM recording | Similar loss of AIMs | [24] | |||
|
|
||||||
| RGS9KO mice | Reserpinized- or haloperidol- treated animals (3 days) | Acute apomorphine | Locomotor activity assay | ↑ Locomotor activity | [24] | |
|
|
||||||
| Acute quinpirole or SKF38393 | Locomotor activity | Similar locomotor activity | [24] | |||
|
|
||||||
| Acute quinpirole or apomorphine | AIM recording | ↑ ↑ AIM score | [24] | |||
|
|
||||||
| Acute SKF38393 | AIM recording | Unchangeable AIM score | [24] | |||
|
|
||||||
| L-dopa-induced dyskinesia in Parkinson’s disease | Rats; unilaterally 6- OHDA-lesioned parkinsonian under L-dopa and benserazide treatment | HSV–RGS9-2 (versus HSV–LacZ) overexpression in the striatum | None | AIM recording | ↓ Dyskinesia | [33] |
|
|
||||||
| Monkeys Macaca fascicularis; MPTP and dyskinetic (parkinsonian under Modoparc- treatment-induced dyskinesia) | HSV–RGS9-2 (versus HSV–LacZ) overexpression in the striatum | None | Parkinsonian monkey rating scale | Similar PD score (antiparkinsonian action of Modopar) | [33] | |
|
|
||||||
| None | Dyskinesia disability scale | ↓ LID | [33] | |||
|
|
||||||
| None | Locomotor activity assay | ↓ Locomotor activity | [33] | |||
|
|
||||||
| Acute ropinirole (D2-receptor and D3-receptor agonist) | Parkinsonian monkey rating scale | ↓ PD score (↓ antiparkinsonian action of Modopar) | [33] | |||
|
|
||||||
| Acute ropinirole | Dyskinesia disability scale | ↓ LID | [33] | |||
|
|
||||||
| Acute ropinirole | Locomotor activity assay | ↓ Locomotor activity | [33] | |||
|
|
||||||
| RGS9KO mice | Unilateral 6-OHDA lesion-induced PD | Chronic L-dopa and benserazide treatment | Rotational and locomotor recording | Similar rotational behavior (antiparkinsonian effect) | [33] | |
|
|
||||||
| Chronic L-dopa and benserazide treatment | AIM rating scale | ↑ Forelimb and axial AIM (dyskinesia) | [33] | |||
|
|
||||||
| Response to psychostimulants | RGS9KO mice | None | Acute amphetamine (low dose) | Locomotor activity | ↑ Locomotor activity | [23] |
|
|
||||||
| Acute amphetamine (high dose) | Locomotor activity | Similar locomotor activity | [23] | |||
|
|
||||||
| Acute cocaine | Locomotor activity | Similar locomotor activity | [23] | |||
|
|
||||||
| Chronic cocaine | Locomotor activity | ↑ Locomotor activity | [23] | |||
|
|
||||||
| Rats | Bilateral HSV-RGS9-2 (versus HSV-LacZ) overexpression in the NAc | Acute cocaine (low dose) | Locomotor activity | ↓ Locomotor activity | [23] | |
|
|
||||||
| Acute cocaine (high dose) | Locomotor activity | Similar locomotor activity | [23] | |||
|
| ||||||
| Antinociception | ||||||
|
|
||||||
| Acute | RGS9KO mice | None | Acute morphine | Hot plate | ↑ Supra-spinal-cord-mediated analgesia | [25] |
|
|
||||||
| Acute morphine | Tail flick | ↑ Spinal-cord-mediated analgesia | [25] | |||
|
|
||||||
| Tolerance | RGS9KO mice | None | Chronic morphine | Hot plate | Delayed tolerance development | [25] |
|
|
||||||
| SNI model of neuropathic pain | Chronic morphine | Von Frey | Delayed tolerance development | Zachariou, V., unpublished | ||
|
| ||||||
| Reward | ||||||
|
|
||||||
| RGS9KO mice | None | Cocaine (low dose) | CPP | ↑ Place conditioning | [23,25] | |
|
|
||||||
| Cocaine (high dose) | CPP | Similar place conditioning | None | |||
|
|
||||||
| Morphine (low dose 0.5–3 mg/kg) | CPP test | ↑ Place conditioning (decreased threshold) | [23,25] | |||
|
| ||||||
| Physical dependence and withdrawal | ||||||
| RGS9KO mice | Morphine dependence (treatment with escalating morphine doses) | Acute naloxone | Withdrawal test | ↑ Opiate withdrawal | [25] | |
Abbreviations: AIM, arbitrary involuntary movement; CPP, conditioned place preference; HSV, herpes simplex virus; LacZ, β galactosidase; LID, L-dopa-induced dyskinesia; 6-OHDA, 6-hydroxydopamine; PD, Parkinson’s disease; SNI, spared nerve injury.
Arrows denote a decrease or increase in behavior compared with control animals.
Modopar is L-dopa plus carbidopa.
The finding that RGS9-2 is highly and specifically expressed in the striatum was a tell-tale sign of a role in dopaminergic signaling and the actions of drugs of abuse. By contrast, RGS9-2 is not the only RGS protein expressed in this region; the smaller proteins RGS2, RGS4 and RGS8 are also present. These RGS proteins cannot substitute for RGS9-2 because introduction of RGS4 into the NAc of RGS9KO does not alter the place-preference response to morphine [25], probably because the protein–protein binding regions of the larger RGS9-2 are important for RGS9–2 actions and because of differences in the relative G-protein specificity. Notably, there is considerable evidence that RGS2, RGS4 and RGS8 in addition to RGS9-2 are modulated by stimulants and opioids in the striatum and other regions [28,29] and can, at least in expression systems, modulate the actions of opioids [30].
The importance of all RGS-related GTPase-accelerating protein (GAP) activity, including RGS9-2 in regulation of physiological processes, can be probed using a functional knockout of GAP activity. This is achieved using mutant Gαi/o proteins that do not bind to the RGS consensus sequence and so are immune to GAP activity, yet retain all other properties of Gαi/o proteins [31]. Using RGS-insensitive Gα proteins, it has been shown in cell systems that endogenous RGS proteins negatively regulate the ability of μ opioids such as morphine to signal to the downstream effectors adenylyl cyclase (AC) and mitogen-activated protein kinase [30]; this confirms a role for RGS proteins in opioid responses. In addition, RGS proteins seem to protect against tolerance of the AC response at the cell level [31]. Also, there is an RGS-mediated inhibition of AC sensitization, which is considered to be a cellular model of withdrawal, in line with the increased morphine withdrawal signs seen in RGS9KO mice [25]. Mice with a genomic knock-in of RGS-insensitive Gαi2, which retain the normal distribution of Gα protein, have been generated to assess physiological roles for RGS proteins acting as GAPs at Gαi2. Preliminary studies with these mice† showed no difference in acute morphine antinociception in the tail withdrawal or hot-plate tests compared with wild-type littermates and no difference in tolerance development. However, naloxone-precipitated withdrawal in the mice expressing RGS-insensitive Gαi2 was reduced compared with the wild-type littermates; this indicates that RGS proteins acting as GAPs at Gαi2 do enhance dependence and/or the withdrawal response itself. This is opposite from the effect seen with RGS9KO mice and indicative of the complexity of this response.
In contrast to actions to limit signaling of opioid receptors to AC and the mitogen-activated protein kinase pathway, RGS proteins seem to have no modulatory effect on the ability of μ opioids to effect the release of calcium from intracellular stores [30] and actually increase intracellular calcium release by dopamine activation of the D2 receptors [32]. This indicates that the release of intracellular calcium might be modulated in the opposite direction to other signaling pathways and that the relative importance of the intracellular pathways will change as the RGS status of the cell changes. This can be particularly important in the face of long-term exposure to drugs of abuse which, as discussed earlier, can change RGS levels.
RGS9-2 overexpression prevents dyskinesias associated with neuroleptic and antiparkinsonian treatments
Several antipsychotic drugs exert their actions partly via interventions in dopamine D2 receptor function. Although it is not known whether RGS9-2 modulates the actions of antipsychotic drugs in the striatum, there is evidence for a possible role of RGS9-2 in neuroleptic-induced extrapyramidal side effects such as dyskinesias. Studies by Kovoor and colleagues [24] showed a role of RGS9-2 in D2-dopamine-receptor-mediated involuntary movements in mice. In these studies, treating RGS9WT and RGS9KO mice with reserpine or haloperidol produced a decrease in spontaneous locomotor activity, and subsequent activation of the D2 receptor by agonists such as quinpirole produced severe abnormal involuntary movements (Table 2). These signs were more prominent in RGS9KO mice, which also showed greater locomotor activation after apomorphine treatment. These data were further supported by cell-culture studies, which demonstrated colocalization of RGS9-2 with dopamine D2 receptors at the cell membrane after agonist treatment [24].
Recently, a study by Gold and colleagues [33] used a primate model to demonstrate that increasing RGS9-2 activity in the striatum can prevent the dyskinesia associated with chronic levodopa (L-dopa) treatment. L-Dopa is widely used in dopamine-replacement therapy in Parkinson’s disease. Chronic L-dopa treatment has several side effects, the most prominent being the development of dyskinesia resulting from imbalanced D1 over D2 receptor signaling. Because earlier data showed that RGS9-2 is a negative regulator of the locomotor actions of D2 but not of D1 agonists, increased RGS9-2 activity could limit the locomotor-activating actions of L-dopa by accelerating signaling termination in D2 neurons. This hypothesis was tested by bilateral infection of a herpes simplex virus expressing RGS9-2 into the striata of 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP)-treated, dyskinetic monkeys. MPTP is used to produce lesions on dopaminergic neurons, similar to those observed in Parkinson’s disease. Animals overexpressing RGS9-2 showed a significantly lower amount of dyskinesia compared with animals infected with a control virus (Table 2). Importantly, increased RGS9-2 activity in the dorsal striatum reduced L-dopa-induced dyskinesia without affecting the antiparkinsonian effects of the drug. These results further support the idea that interventions in receptor-subtype-specific-signaling events can be used to limit the undesired effects of a drug.
Additional evidence of the regulation of D2-receptor function by RGS9-2 derives from recordings from striatal slices expressing different forms of RGS9.
The study by Cabrera-Vera and colleagues [34] provided information on the actions of RGS9-2 at the level of a single neuron. Recordings from striatal cholinergic interneurons (which also express RGS9-2 and Gβ5) indicate that overexpression of RGS9-2 inhibits D2 receptor modulation of calcium Cav.v2 channels and enhances basal Ca2+ currents [34] (Table 2).
Conclusions and future perspectives
The determinants of the selectivity of action of RGS proteins are not only their differential tissue distribution, as with striatal expression of RGS9-2, but also their selectivity for particular receptor types and levels of receptor, cognate G proteins and effectors [35]. There are several strategies via which RGS proteins can be used as therapeutic targets. If different effects of drugs are modulated by specific RGS proteins then it should be possible to target these selectively and enhance desirable actions (e.g. analgesia) at the expense of unwanted actions, even though all are mediated by a single, in this case, μ-opioid receptor. This approach can be particularly effective with partial-agonist analgesics that already have a safer side-effect profile. Finally, gene-therapy approaches, using adeno-associated viruses or other viral vectors to overexpress an RGS protein in particular brain regions, could also provide a therapeutic approach, especially in case of neurodegenerative disorders. RGS proteins interact with other signal-transduction molecules via protein–protein interactions, and designing inhibitors of such protein–protein interaction has proved challenging. Although very few reports have been published on compounds that selectively target RGS proteins, a peptide inhibitor, YJ34 [36], in addition to a small molecular inhibitor, CGC-4986 [37], have been described, indicating that it is possible to overcome this hurdle. Alternatives are to target sites on the RGS proteins that are not directly involved in protein–protein interactions but can control RGS activity such as the phosphatidylinositol (3,4,5)-trisphosphate binding site within the RGS consensus domain [38]. Another major challenge is to design compounds that can cross the blood–brain barrier and act on particular neuronal sub-populations. Thus, understanding of the cell-type-specific actions of RGS9-2 in the striatum is crucial for the development of new therapeutic strategies and for the generation of tools targeting this protein.
Acknowledgments
Supported by DA04087 (J.R.T.) and by the Greek Secretariat for Research and Technology (www.gsrt.gr; PENED03; V.Z.).
Footnotes
Terzi D. and Zachariou V. (2008) RGS9 modulates analgesic effects of μ, δ and κ opioid receptor agonists, and depression like behaviors in a mouse model of neuropathic pain [abstract]. Soc. Neurosci. Abstr. 2008, Abstract 759.
Talbot, J.N. et al. (2008) Genetic deletion of regulators of G protein signaling protein activity enhances buprenorphine antincoiception while limiting withdrawal behaviors associated with chronic administration [abstract]. Experimental Biology 2008, San Diego, Abstact 907.
References
- 1.Robbins TW, et al. Limbic-striatal interactions in reward-related processes. Neurosci Biobehav Rev. 1989;13:155–162. doi: 10.1016/s0149-7634(89)80025-9. [DOI] [PubMed] [Google Scholar]
- 2.Picconi B, et al. Pathological synaptic plasticity in the striatum: implications for Parkinson’s disease. Neurotoxicology. 2005;26:779–783. doi: 10.1016/j.neuro.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 3.Nestler EJ, Carlezon WA., Jr The mesolimbic dopamine reward circuit in depression. Biol Psychiatry. 2006;59:1151–1159. doi: 10.1016/j.biopsych.2005.09.018. [DOI] [PubMed] [Google Scholar]
- 4.Everitt BJ, Robbins TW. Neural systems of reinforcement for drug addiction: from actions to habits to compulsion. Nat Neurosci. 2005;8:1481–1489. doi: 10.1038/nn1579. [DOI] [PubMed] [Google Scholar]
- 5.Hepler JR, Gilman AG. Trends Biochem Sci. 1992;17:383–387. doi: 10.1016/0968-0004(92)90005-t. [DOI] [PubMed] [Google Scholar]
- 6.Dohlman HG, et al. Sst2, a negative regulator of pheromone signaling in the yeast Saccharomyces cerevisiae: expression, localization, and genetic interaction and physical association with Gpa1 (the G-protein α subunit) Mol Cell Biol. 1996;16:5194–5209. doi: 10.1128/mcb.16.9.5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ross EM, Wilkie TM. GTPase-activating proteins for heterotrimeric G proteins: regulators of G protein signaling (RGS) and RGS-like proteins. Annu Rev Biochem. 2000;69:795–827. doi: 10.1146/annurev.biochem.69.1.795. [DOI] [PubMed] [Google Scholar]
- 8.Hepler JR, et al. RGS and GAIP are GTPase activating proteins for Gqα and block activation of phospholipase Cβ by γ-thio-GTP-Gq-α. Proc Natl Acad Sci U S A. 1997;94:428–432. doi: 10.1073/pnas.94.2.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Richman RW, et al. GS12 interacts with the SNARE-binding region of the Cav2.2 calcium channel. J Biol Chem. 2005;280:1521–1528. doi: 10.1074/jbc.M406607200. [DOI] [PubMed] [Google Scholar]
- 10.Gold SJ, et al. Regulators of G protein signaling (RGS) proteins: region-specific expression of nine subtypes in rat brain. J Neurosci. 1997;17:8024–8037. doi: 10.1523/JNEUROSCI.17-20-08024.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Neubig RR, Siderovski DP. Regulators of G-protein signaling as new central nervous system drug targets. Nat Rev Drug Discov. 2002;1:187–197. doi: 10.1038/nrd747. [DOI] [PubMed] [Google Scholar]
- 12.Labouèbe G, et al. RGS2 modulates coupling between GABAB receptors and GIRK channels in dopamine neurons of the ventral tegmental area. Nat Neurosci. 2007;10:1559–1568. doi: 10.1038/nn2006. [DOI] [PubMed] [Google Scholar]
- 13.Charlton JJ, et al. Multiple actions of spinophilin modulate mu opioid receptor function. Neuron. 2008;58:238–247. doi: 10.1016/j.neuron.2008.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chase DL, et al. Mechanism of extrasynaptic dopamine signaling in Caenorhabditis elegans. Nat Neurosci. 2004;7:1096–1103. doi: 10.1038/nn1316. [DOI] [PubMed] [Google Scholar]
- 15.Hooks SB, et al. A role of RGS proteins in drug addiction. Biochem Pharmacol. 2008;75:76–84. doi: 10.1016/j.bcp.2007.07.045. [DOI] [PubMed] [Google Scholar]
- 16.Levitt P, et al. Making the case for a candidate vulnerability gene in schizophrenia: convergent evidence for regulator of G-protein signaling 4 (RGS4) Biol Psychiatry. 2005;60:534–537. doi: 10.1016/j.biopsych.2006.04.028. [DOI] [PubMed] [Google Scholar]
- 17.Nishiguchi KM, et al. Defects in RGS9 or its anchor protein R9AP in patients with slow photoreceptor deactivation. Nature. 2004;427:75–78. doi: 10.1038/nature02170. [DOI] [PubMed] [Google Scholar]
- 18.Martemyanov KA, et al. R7BP, a novel neuronal protein interacting with RGS proteins of the R7 family. J Biol Chem. 2005;280:5133–5136. doi: 10.1074/jbc.C400596200. [DOI] [PubMed] [Google Scholar]
- 19.Witherow DS, et al. Complexes of the G protein subunit Gβ5 with the regulators of G protein signaling RGS7 and RGS9. Characterization in native tissues and in transfected cells. J Biol Chem. 2000;275:24872–24880. doi: 10.1074/jbc.M001535200. [DOI] [PubMed] [Google Scholar]
- 20.Chen CK, et al. Instability of GGL domain-containing RGS proteins in mice lacking the G protein β-subunit Gβ5. Proc Natl Acad Sci U S A. 2003;100:6604–6609. doi: 10.1073/pnas.0631825100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hooks SB, et al. RGS6, RGS7, RGS9, and RGS11 stimulate GTPase activity of Gi family G proteins with differential selectivity and maximal activity. J Biol Chem. 2003;278:10087–10093. doi: 10.1074/jbc.M211382200. [DOI] [PubMed] [Google Scholar]
- 22.Rahman Z, et al. Cloning and characterization of RGS9-2, a striatal enriched alternatively sliced product of the RGS9 gene. J Neurosci. 1999;19:2016–2026. doi: 10.1523/JNEUROSCI.19-06-02016.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rahman Z, et al. RGS9 modulates dopamine signaling in the basal ganglia. Neuron. 2003;38:941–952. doi: 10.1016/s0896-6273(03)00321-0. [DOI] [PubMed] [Google Scholar]
- 24.Kovoor A, et al. D2 dopamine receptors colocalize regulator of G protein signaling 9-2 (RGS9-2) via the RGS9 DEP domain, and RGS9 knockout mice develop dyskinesias associated with dopamine pathways. J Neurosci. 2005;25:2157–2165. doi: 10.1523/JNEUROSCI.2840-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zachariou V, et al. Essential role for RGS9 in opiate action. Proc Natl Acad Sci U S A. 2003;100:13656–13661. doi: 10.1073/pnas.2232594100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blundell J, et al. Motor coordination deficits in mice lacking RGS9. Brain Res. 2008;1190:78–85. doi: 10.1016/j.brainres.2007.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Psifogeorgou K, et al. RGS9 is a negative modulator of μ opioid receptor function. J Neurochem. 2007;103:617–625. doi: 10.1111/j.1471-4159.2007.04812.x. [DOI] [PubMed] [Google Scholar]
- 28.Gold SJ, et al. Regulation of RGS proteins by chronic morphine in rat locus coeruleus. Eur J Neurosci. 2003;17:971–980. doi: 10.1046/j.1460-9568.2003.02529.x. [DOI] [PubMed] [Google Scholar]
- 29.Schwendt M, et al. Acute amphetamine down-regulates RGS4 mRNA and protein in rat forebrain: distinct roles of D1 and D2 dopamine receptors. J Neurochem. 2006;96:1606–1615. doi: 10.1111/j.1471-4159.2006.03669.x. [DOI] [PubMed] [Google Scholar]
- 30.Clark MJ, et al. Endogenous RGS protein action modulates μ-opioid signaling through Gαo. Effects on adenylyl cyclase, extracellular signal-regulated kinases, and intracellular calcium pathways. J Biol Chem. 2003;278:9418–9425. doi: 10.1074/jbc.M208885200. [DOI] [PubMed] [Google Scholar]
- 31.Clark MJ, Traynor JR. Endogenous regulator of G protein signaling proteins reduce μ-opioid receptor desensitization and down-regulation and adenylyl cyclase tolerance in C6 cells. J Pharmacol Exp Ther. 2005;312:809–815. doi: 10.1124/jpet.104.074641. [DOI] [PubMed] [Google Scholar]
- 32.Boutet-Robinet EA, et al. Endogenous RGS proteins facilitate dopamine D2 receptor coupling to Gαo proteins and Ca2+ responses in CHO-K1 cells. FEBS Lett. 2003;533:67–71. doi: 10.1016/s0014-5793(02)03753-5. [DOI] [PubMed] [Google Scholar]
- 33.Gold SJ, et al. RGS9-2 negatively modulates L-3, 4-dihydroxyphenylalanine-induced dyskinesia in experimental Parkinson’s disease. J Neurosci. 2007;27:14338–14348. doi: 10.1523/JNEUROSCI.4223-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cabrera-Vera TM, et al. RGS9-2 modulates D2 dopamine receptor-mediated Ca2_ channel inhibition in rat striatal cholinergic interneurons. Proc Natl Acad Sci U S A. 2004;101:16339–16344. doi: 10.1073/pnas.0407416101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Clark MJ, et al. Endogenous regulators of G protein signaling differentially modulate full and partial mu-opioid agonists at adenylyl cyclase as predicted by a collision coupling model. Mol Pharmacol. 2008;73:1538–1548. doi: 10.1124/mol.107.043547. [DOI] [PubMed] [Google Scholar]
- 36.Jin Y, et al. Structure-based design, synthesis, and activity of peptide inhibitors of RGS4 GAP activity. Methods Enzymol. 2004;389:266–277. doi: 10.1016/S0076-6879(04)89016-5. [DOI] [PubMed] [Google Scholar]
- 37.Roman DL, et al. Identification of small-molecule inhibitors of RGS4 using a high-throughput flow cytometry protein interaction assay. Mol Pharmacol. 2007;71:169–175. doi: 10.1124/mol.106.028670. [DOI] [PubMed] [Google Scholar]
- 38.Traynor JR, Neubig RR. Regulators of G protein signaling and drugs of abuse. Mol Interv. 2005;5:30–41. doi: 10.1124/mi.5.1.7. [DOI] [PubMed] [Google Scholar]