Abstract
Amyotrophic Lateral Sclerosis (ALS) is a progressive and disabling neurodegenerative disorder characterized by upper and lower motor neuron loss, leading to respiratory insufficiency and death after 3-5 years. Riluzole is currently the only FDA approved drug for ALS, but it has only modest effects on survival. The majority of ALS cases are sporadic and probably associated to a multifactorial etiology. With the completion of genome sequencing in humans and model organisms, together with the advent of DNA microarray technology, the transcriptional cascades and networks underlying neurodegeneration in ALS are being elucidated providing new potential pharmacological targets. The main challenge now is the effective screening of the myriad of targets to identify those with the most therapeutic utility. The present review will illustrate how the identification, prioritization and validation of preclinical therapeutics can be achieved through genomic analysis of critical pathways and networks deregulated in ALS pathology.
Keywords: ALS, drug, pathway, pharmacogenomics, networks, target.
INTRODUCTION
Amyotrophic Lateral Sclerosis (ALS) is a progressive, disabling neurodegenerative disorder characterized by upper and lower motor neuron loss, leading to respiratory insufficiency and death after 3-5 years [1, 2]. The incidence of ALS ranges from 1.7 to 2.3 cases per 100,000 population per year world-wide [3]. Currently, ALS is an incurable disease and the only FDA approved drug, Riluzole, has very modest efficacy on survival [4].
Despite intensive research, knowledge of the pathogenetic mechanisms and precise genetic causes of ALS remains incomplete. Although most cases of ALS are sporadic (SALS), about 10% are familial (FALS), mostly with autosomal dominant inheritance [5]. In 3-7% of all ALS and 20% of FALS cases, different mutations in the gene encoding copper–zinc superoxide dismutase (SOD1) have been found [6]. In addition to SOD1, several mutations in other genes, including Senataxin (SETX) [7], Vesicle-associated associated protein B (VAPB) [8], and Alsin (ALS2) [9, 10], Spatacsin (SPG11) [11], Angiogenin (ANG) [12], PI(3,5)P(2)5-phosphatase (Fig. 4) [13], and Optineurin (OPTN) [14] have been identified as causative for classical FALS. The etiology of SALS is still unknown but it is now widely accepted that SALS is a multi-factorial complex disease, which is attributable to and influenced by the interaction of environmental factors with multiple genes, such as those encoding for the heavy neurofilament subunit, peripherin, dynactin, and FLJ10986 [15]. An exciting step forward in ALS genetics is represented by the recent discovery of mutations in TDP-43 (encoded by TARDBP) [16] and the related RNA-binding protein fused in sarcoma/translocated in liposarcoma (FUS) [17] in familial and sporadic cases that has shifted the focus of much research on RNA metabolism, and implicated abnormal RNA processing in ALS pathogenesis. Further advances in ALS etiology have also been obtained through the use of the new next-generation sequencing technology that allows to rapidly screen common genetic variation across the human genome [18]. A two-stage genome-wide association study (GWAS), which included data from nearly 20000 patients with sporadic ALS, has identified mutations in the UNC13A (unc-13 homolog A) gene and found a significant association with the 9p21 chromosomal locus [19]. Importantly, the same chromosomal region has been confirmed in large independent GWASs of both ALS and Frontotemporal dementia (FTD), implicating the genetic defect at chromosome 9p in sporadic forms of both diseases [19-22]. Furthermore, by the same approach an expanded hexanucleotide repeat in a noncoding region of chromosome 9 open reading frame 72 (C9ORF72) has been identified as the most common cause of familial ALS, FTD and ALS-FTD forms [23, 24]. Despite common genetic variants are beginning to be unequivocally linked to ALS, to date its pathogenesis is not clearly understood. Additional works therefore are needed to better understand and effectively treat this disease.
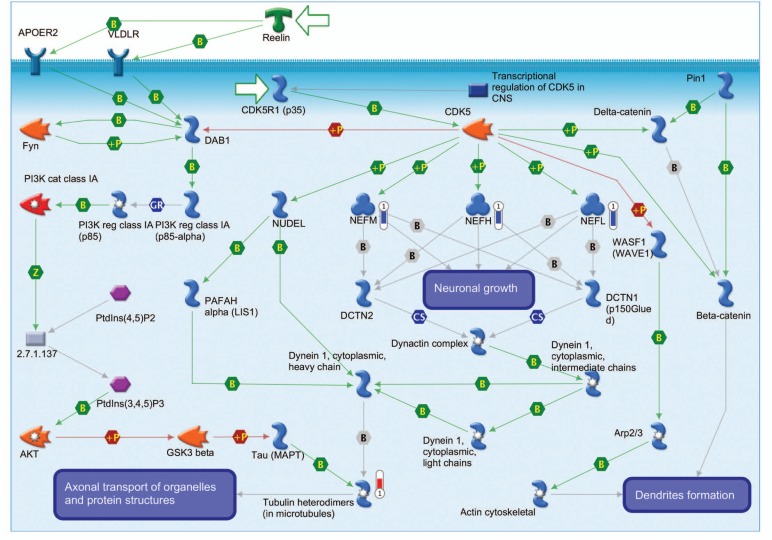
Fig. (4).
Cytoskeleton remodeling signaling, an example of a differentially regulated pathway in the motor cortex of SALS. Thermometers labeled with (1) or (2) indicate expression levels in motor cortex and spinal cord, respectively. Downward thermometers have blue color and indicate down-regulated expression, whereas upward thermometers have red color and indicate up-regulated expression. The mechanism of physical interaction is indicated: B, binding; +P, phosphorylation; T, transformation; TR, transcription regulation; Z, catalysis.
While the deep genome sequencing technologies are rapidly emerging as a useful method for the discovery of ALS susceptibility and causative genes, DNA-microarray is currently the most widely high throughput technology used to clarify the ALS pathogenic mechanisms. In the last ten years, genome-wide expression analysis by DNA microarray technology has been conducted on various tissues from rodent models [25-33] and ALS patients [34-36]. Our research group, in particular, has identified genes de-regulated in the motor cortex of patients with sporadic ALS, and interpreted the role of individual candidate genes in a framework of differentially expressed pathways [36]. Genomic-based studies are beginning to elucidate the transcriptional cascades and networks underlying neurodegeneration in ALS, drawing a precise molecular portrait of the pathology [37]. In addition to providing an unprecedented experimental opportunity to investigate ALS disease, gene expression profiling studies have also allowed the identification of new potential pharmacological targets. The main challenge is now the effective screening of the potential targets to identify those with the most therapeutic utility.
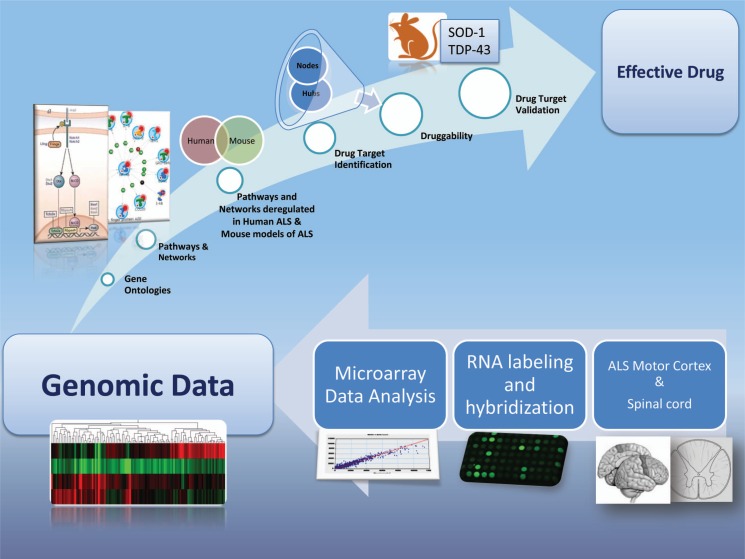
In the present review, we will illustrate how the identification, prioritization and validation of preclinical therapeutics can be achieved through microarray-based transcriptomic analysis of critical pathways and networks deregulated in ALS pathology (Fig. 1).
Fig. (1).
Genomic approach for drug discovery in ALS. A drug discovery pipeline starting from microarray analysis moves through drug targets identification and ends with their validation in animal models of ALS.
DIFFERENTIALLY EXPRESSED GENES RELEVANT TO ALS
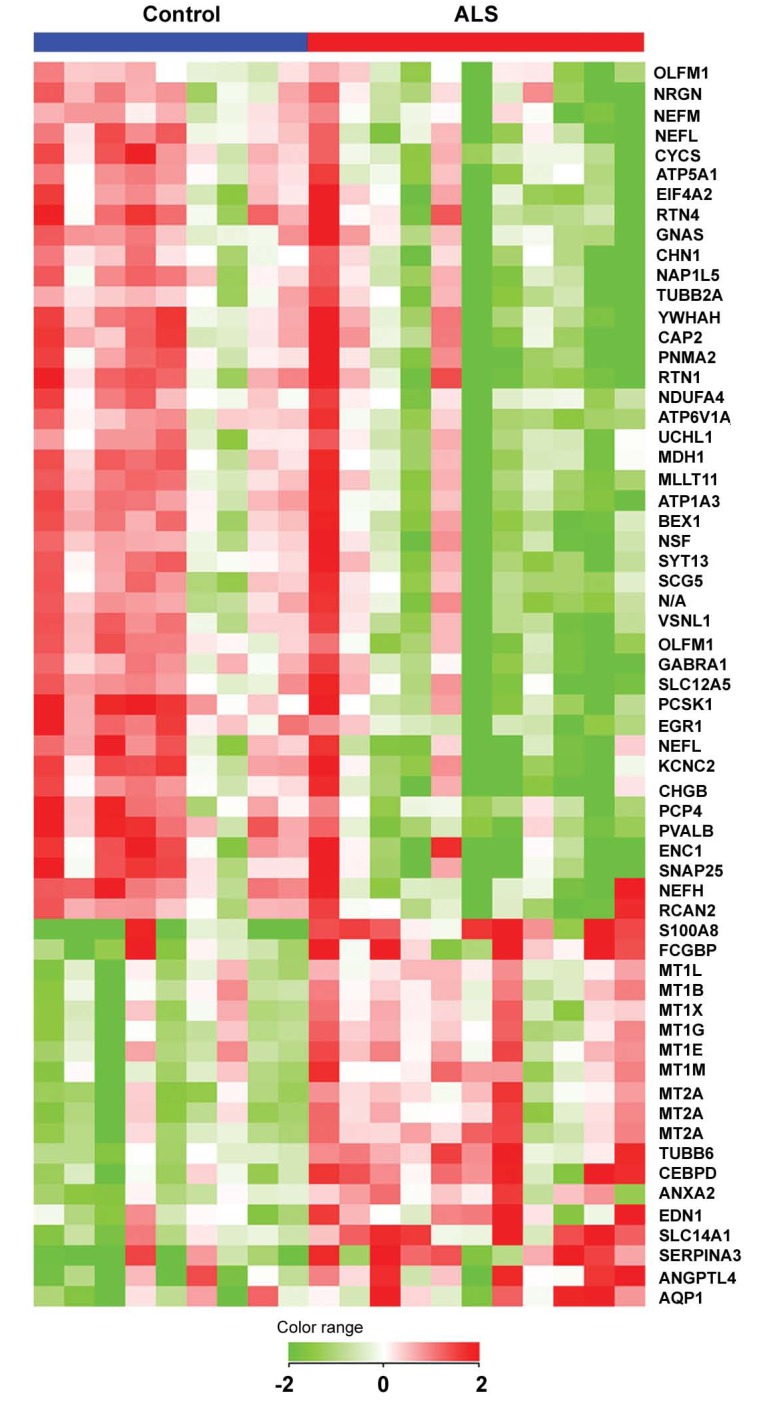
As previously discussed, the etio-pathogenesis of sporadic ALS is largely unknown but it is now widely accepted that SALS is a multifactorial complex disease, which is attributable and influenced by the interaction of environmental factors and lifestyles with multiple genes. The sequencing of the human genome, and the development of high throughput technologies, offer today an unprecedented experimental opportunity to investigate ALS disease through a genomic approach. To illustrate this approach, we will refer to our previous published study where we examined whole genome expression profiles of motor cortex in control and sporadic ALS patient [36]. This screening revealed the differential expression of 57 genes (40 genes showing down-regulation and 17 up-regulation) (Fig. 2), which may be involved in ALS neurodegeneration and offer potential pharmacological targets. To exemplify the use of single genes as potential pharmacological targets, below we will discuss the down regulation of four genes whose encoding proteins are involved with ion homeostasis and excitotoxicity: GABA-A receptor alpha1-subunit (GABRA1), ATPase - Na+/K+ transporting - alpha 3 polypeptide (ATP1A3), potassium voltage-gated channel subfamily C member 2 (KCNC2), and solute carrier family 12 (potassium/chloride transporter) - member 5 (SLC12A5). For a complete description of all the differentially expressed genes, the reader is referred to our previous study [36]. Reduced level of GABRA1 was previously demonstrated in human motor cortex of ALS patients and was associated with heightened excitability [38]. Mutations of the plasma-membrane Na+/K+ pump ATP1A3 have been linked to rapid-onset dystonia parkinsonism [39] and its expression is reduced in SOD1(G93A) FALS mice [40] and in motoneurons of Wobbler mice [41]. KCNC2 regulates the voltage-dependent potassium ion permeability of excitable membranes and its decreased expression may lead to decreased potassium conductance and delayed repolarization of axons [42]. SLC12A5 encodes the potassium-chloride cotransporter KCC2, a neuronal isoform of the potassium-chloride cotransporter family [43]. During early development, increased expression of SLC12A5 lowers the intra-neuronal chloride concentration below its electrochemical equilibrium and allows GABA to act as an inhibitory neurotransmitter [44]. Conversely, a switch of GABA action from inhibitory to excitatory has been proposed as a mechanism contributing to excitotoxicity in injured neurons. Indeed, down-regulation of SLC12A5 expression together with GABAA receptor-mediated excitation occurs after axonal or spinal cord injury [45, 46], and mouse SLC12A5 knockouts suffer severe motor deficits and immediate postnatal death by asphyxiation [47].
Fig. (2).
Hierarchical cluster of genes differentially expressed in motor cortex of SALS subjects. 57 of 19,431 quality-filtered genes (0.3%), represented by 61 probes, are differentially expressed, with each row in the matrix representing a single probe and each column a subject. Normalized expression levels are represented by the color of the corresponding cell, relative to the median abundance of each gene for each subject (see scale). Genes are named using their UniGene symbol and arranged in a hierarchical cluster (standard correlation) based on their expression patterns, combined with a dendrogram whose branch lengths reflect the relatedness of expression patterns.
As we have seen in the previous paragraph, microarray analysis allows identifying genes that are significantly differentially expressed in ALS. Although, some of their encoded proteins may be potential pharmacological targets, differentially expressed genes represent only the tip of the “iceberg” of a genomic analysis. When genes are analyzed individually, small changes in expression may not pass stringent statistical cut off. Those small changes; however, may show a statistical significance when analyzed, for example, in the context of a pathway or a network. In the following sections, we will illustrate how these types of analyses are of fundamental help to extract more knowledge and discover more complex relationships from genomic data.
GENE ONTOLOGIES, PATHWAYS AND PROTEIN INTERACTING NETWORKS: THEIR IMPLICATION FOR DRUG TARGET DISCOVERY IN ALS
Cellular processes depend on the activity of an integrated network of genes and their encoded proteins, which almost never work alone but interact with one another in highly structured and incredibly complex ways. In this integrated network it is not important the activity of the single gene and their encoded protein, but the entire components and their interactions, a concept that can be summarized in: “The whole is greater than the sum of its parts”. Thus, genes do not act by themselves, but they function in gene networks and molecular pathways and their effects are not independent but often modified by one or several other genes (epistasis) [48]. Gene ontology enrichment and protein interacting networks of microarray data allow us to look at the “whole” instead of the “single parts”.
Gene Ontology Enrichment
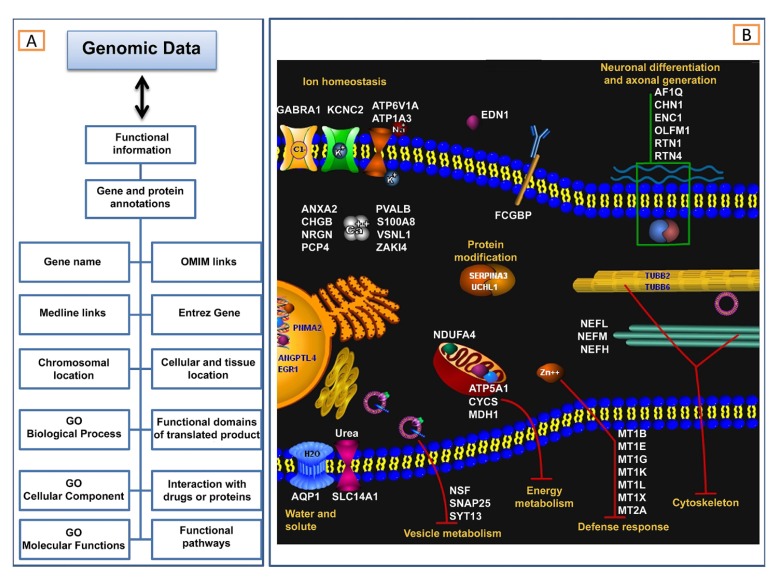
A gene or its encoded protein has not only a name/symbol or an expression value, but also several ontologies or functional annotations (Fig. 3A).
Fig. (3).
Gene ontologies. (A) Correlations between microarray data and functional information. Complex correlations between gene expression profiles and functional annotations are needed to extract more knowledge. Examples of functional information are listed on the left side of the figure and include gene or protein annotations. (B) GO analysis of genes differently expressed in cortex of sporadic ALS patients reveals the involvement of specific cellular processes and sub-cellular compartments.
Common functional annotations are those listed in the Gene Ontology (GO) database (www.geneontology.org), a controlled vocabulary of terms that describes the roles of genes and proteins in all organisms [49]. GO is comprised of three independent ontologies: 1) biological process describes biological goals accomplished by one or more ordered assemblies of molecular functions; 2) cellular component describes locations, at the levels of sub cellular structures and macromolecular complexes; 3) molecular function describes activities, such as catalytic or binding activities, at the molecular level. Biological process, molecular function and cellular component are all attributes of genes, gene products or gene-product groups and each of these may be assigned independently. The relationships between a gene product (or a gene-product group) to biological process, molecular function and cellular component are one-to-many, reflecting the biological reality that a particular protein may function in several processes, contain domains that carry out diverse molecular functions, and participate in multiple alternative interactions with other proteins, organelles or locations in the cell. Through the use of GO terms, a number of software tools (www.geneontology.org/GO.tools.shtml) are able to perform gene ontology enrichment analysis of high-throughput experimental results, such as gene expression microarray data, and discover statistically significantly enriched GO terms among a given gene list. An example of gene ontology enrichment based on GO terms is shown in Fig. (3B), where GO analysis of genes differently expressed in cortex of sporadic ALS patients reveals the involvement of specific cellular processes and sub-cellular compartments.
Pathways Analysis
In addition to the GO terms described above, many other annotations are nowadays linked to a specific gene/protein. Examples of these are the associated disease (OMIM Links), publications (Medline links), chromosomal location, interacting drug, functional domain, and functional pathway (Fig. 3A). This last, in particular, represents a set of consecutive signals or metabolic transformations that have been confirmed as a whole by experimental data. Thousand of pathways are nowadays available (for a list of biological pathway related resources see: www.pathguide.org) and different informatics tools have been developed that enable to analyze gene expression changes in the context of pathways. Below we describe some of these public and private resources. The Kyoto Encyclopedia of Genes and Genomes (KEGG) [50] is a free resource that contains a comprehensive collection of databases for genes, pathways and ligands for several organisms, together with web-accessible tools for the retrieval of pathways and the annotation of gene lists. The Gene Map Annotator and Pathway Profiler (GenMAPP) [51] are freely available programs for viewing and analyzing gene expression data in the context of biological pathways. Examples of private resources include MetaCore (www.genego.com), Ingenuity Pathways Analysis (www.ingenuity.com), Pathway Assist (www.ariadnegenomics.com) and GeneSpring (www.agilent.com).
Pathway analysis of genes differentially expressed in motor cortex of ALS patients has been described in more detail in our previous study [36]. In the following paragraph and in Fig. (4), we describe an example of a pathway differentially affected in ALS, the Cytoskeleton remodeling signaling.
The cytoskeleton is critical for neuronal maintenance and plasticity, neurite outgrowth, axonal calibre and transport. As illustrated in Fig. (4), our pathway-based analysis in motor cortex of SALS patient reveals the alteration of two major components of the neuronal cytoskeleton in the ALS motor cortex, showing a general down-regulation of microtubules and deregulation of genes encoding tubulin proteins (Tubulin heterodimers), as well as decreased expression of all three neurofilament subunits (NEFM, NEFL, NEFH). A depletion of microtubules and neurofilaments has deleterious effects on motoneurons, according to our understanding of their role in ALS pathogenesis [52]. Impaired microtubule-based axonal transport causes related motoneuropathies and is the earliest detectable, presymptomatic abnormality in SOD1-mutant FALS mice. In addition to this, defects in microtubule-associated motor proteins cause ALS-related human motoneuropathies, such as Charcot-Marie-Tooth disease [53] and hereditary spastic paraplegia [54], and are responsible for ALS phenotypes in Drosophila [55] and mouse [56]. The role of neurofilaments (NFs) in ALS is still controversial, despite a substantial body of research addressing the subject [52]. Deletion of the NEFL subunits in the SOD1 G85R mouse model is accompanied by preferential increase of the NEFH and NEFM subunits in the motor neuron cell bodies and reduction of these subunits in the axons, with an overall significant delay in the onset and progression of clinical disease [57]. Over-expression of the NEFH subunit has similar effects [58], prompting the hypothesis that NFs primarily act as an abundant buffer for otherwise deleterious processes, such as offering phosphorylation sites for deregulated intracellular kinases, or reducing the burden of axonal transport [52, 59, 60]. Furthermore, decreasing the axonal burden of neurofilaments may protect motor neurons, at least in part, by enhancing axonal transport, a hypothesis supported by the observation of defects in slow axonal transport in presymptomatic mutant SOD1 mice [61]. The down-regulation of all three NF subunits observed in our study would be detrimental according to either interpretation of NF involvement in ALS pathology, first by interfering with their stoichiometric balance, and second by depleting their availability as a potential buffer for aberrant enzymatic activities.
The above described example, other than visualizing gene expression changes in the context of a pre-drawn pathway, allows us to obtain a more complete and comprehensive view of ALS pathogenic mechanisms, providing a rational approach for drug target selection in ALS. Topics relating to drug selection, druggability and drug target validation will be discussed in the next paragraphs.
Network Analysis and Druggability of Nodes and Hubs
Gene and protein-protein interactions are fundamental to all biological processes. A comprehensive determination of all interactions (interactome) [62] that can take place in an organism provides a framework for understanding biology as an integrated system (system biology) [63]. Network analysis has become a fundamental component of systems biology. Because such analysis provides a unifying language to describe relations within complex systems, it has assumed an increasingly significant role in understanding physiological and pathophysiological functions. Discovering the dynamic nature of cellular networks has relevance to human health, since defects in signaling and regulatory pathways are associated with many diseases, such as neurodegenerative disorders [64, 65]. In view of the gene and protein signaling networks associated to a complex disorder such as ALS, we may need to rethink our strategies for drug development, targeting ALS pathogenesis as a system rather than on the level of the single protein molecule. The current trend in pharmaceutical drug development is characterized by a re-evaluation of the “one disease-one drug target” paradigm that has dominated thinking in the pharmaceutical industry for the last few decades [66]. It is now widely accepted that many compounds do not exert their effects through a single target, instead they have multiple targets. In fact, drugs produce perturbations in a molecular system and very often modulate multiple target proteins.
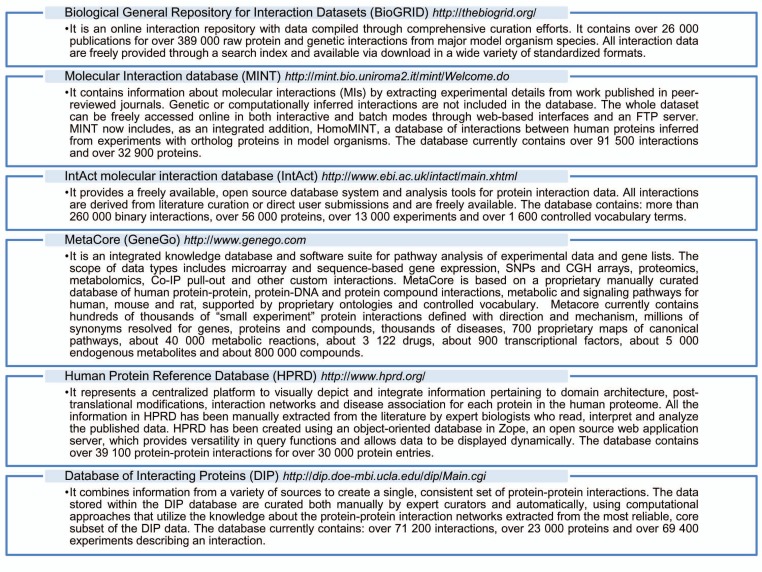
A network can be graphically represented by nodes (genes, proteins) connected by edges (nature of the interaction) and hubs (nodes connected to relatively many other nodes). There are several computational tools and protein-protein interaction database that can help to construct networks from high-dimensional biological datasets, such as microarray data. Some of these computational tools/protein-protein interaction databases are reported in Fig. (5) [2, 67-71]. In the next paragraph, we describe an example of a network of proteins encoded by differently expressed genes in SALS motor cortex, the Metallothioneins network.
Fig. (5).
Several computational tools/protein-protein interaction databases.
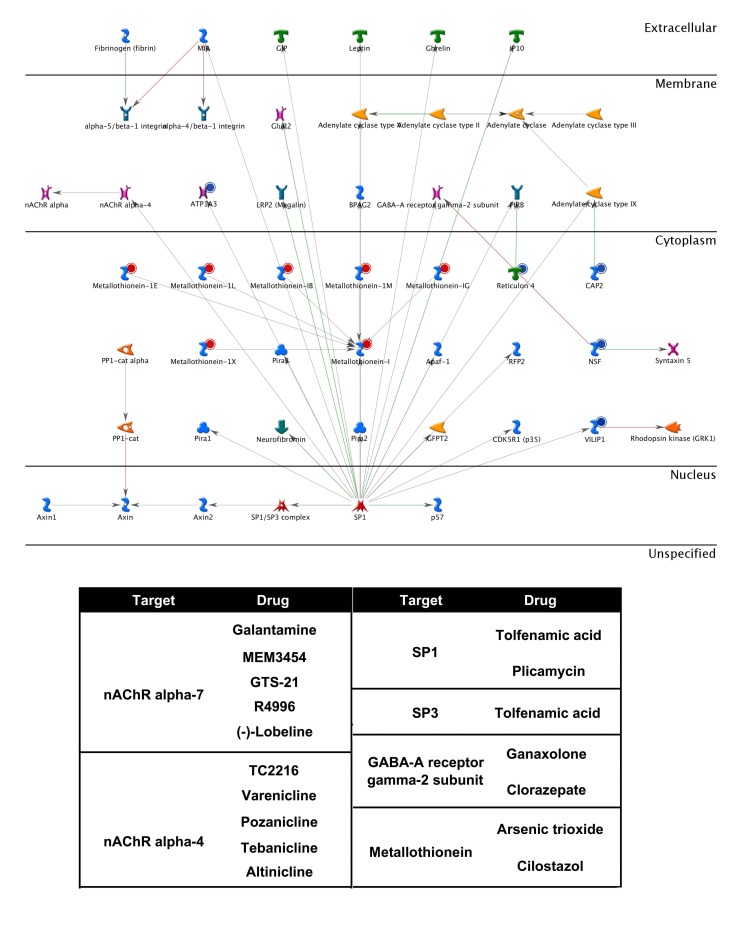
As illustrated in Fig. (6), our previous genomic analysis in SALS patients reveals the coordinated and massive up-regulation of seven different metallothioneins (MT1E, MT1G, MT1L, MT1M, MT1X, MT1B and MT1). Metallothioneins (MTs) are low molecular weight, metal-binding proteins that act as important regulators of metal homeostasis (zinc and copper homeostasis), and as a source of zinc for incorporation into proteins, including zinc-dependent transcription factors [72]. Metallothioneins of the MT1 and MT2 families are able to prevent zinc deficiency in vivo [73], and have also been proposed to function as detoxifiers of heavy metals (zinc, cadmium, inorganic mercury and selenium) and free radicals [74, 75]. In addition to this, MTs seem to have a protective role in the CNS. Indeed, MT-1 and -2 increase in spinal cord of ALS patients and in transgenic mutant-SOD1 mice, where their experimental reduction significantly reduces survival. Importantly, endogenous MT1 and MT2 are undetectable in pure motoneuron cultures, which can be protected against oxidative stress by experimental MT1 over-expression [76], indicating the importance of metallothionein-mediated protection that is normally provided by astrocytes. Furthermore, exogenous MT1 and MT2 uptake promotes axon regeneration in vitro and in vivo, suggesting their potential as therapeutic agents [77].
Fig. (6).
The Metallothioneins signaling network. Genes differentially expressed in motor cortex of sporadic ALS patients were uploaded into the MetaCore web portal (GeneGo) and their translated products (objects) used as the input list for generation of biological networks by the Analyze network algorithm with default settings. Up-regulated genes are marked with red circles, down regulated with blue circles; the green and red lines represent the activation or inhibition of the pathway, respectively. The network algorithm starts with building a “large network” by expanding the initial list of objects. Then, the large network is “cut” into smaller sub-networks highly saturated with objects of the input list, ranked by P-value and interpreted in terms of Gene Ontologies. The different objects (proteins, enzymes, receptors, channels, ligands) identified as pharmacological targets in the biological network, together with their drugs, are reported in the table at the bottom of the figure.
Once identified, deregulated networks nodes and hubs can be topologically investigated for their gene expression pattern and their “druggability”, or else the ability of these objects to be targets for drugs currently on the market or molecules in the development pipeline [78-80]. Fig. (7) shows some bioinformatics and cheminformatics resources [50, 69, 81-85] that combine detailed drug data with comprehensive drug target information and allow investigating the targets druggability. As represented on Fig. (6) for the Metallothioneins network, a list of interacting drug-targets can be identified trough the use of these resources. Some drugs directly interact with their targets regulating their activities, whereas other drugs may interfere with target expression levels. For instance, the Cilostazol, an antiplatelet drug used for the treatment of intermittent claudication, may significantly increase MT protein levels in human neurons as an anti-oxidative effecter molecule [86, 87].
Fig. (7).
Example of some drug-target repositories.
DRUG TARGET VALIDATION
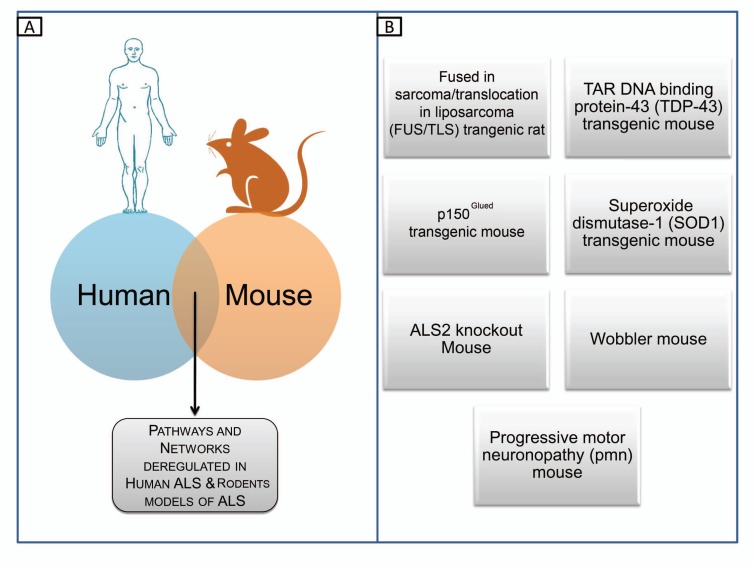
Once drug targets are established, they need to be validated. As we will discuss in the next section, this target validation consists in the accurate evaluation that a specific target is critically involved in a disease process and that modulates the target, is likely to have a desired therapeutic effect. The drug validation process relies on the use of different animal species as models of safety and efficacy before a new compound is administered to humans. Target validation studies are often conducted in mice due to their relatively small size, short generation times and the existence of capabilities such as gene knockout technology, which can be seen as analogous to antagonist treatment. All these experiments assume that modulation of a drug target will have a similar effect on the model species, as it would do on humans. Indeed, since the biology of the drug target differs across species and animal studies do not always translate successfully to humans, attrition rates in drug discovery remain very high [88]. In ALS, target validation is very often performed in rodent models that, as anticipated in the introduction, are monogenic and do not fully represent the complexity seen in human pathology. SALS is a polygenic and multifactorial disease and the utility of these animal models in the preclinical phase of pharmacological trials has been doubted. With these limitations, however, the use of these models is inevitable. Some established rodent models of ALS are listed in Fig. (8). Among these, the recent models based on (mutant) TDP-43 and FUS/TLS lack specificity in relation to the mutation and/or the cell type affected and are not yet used for routine drug screening [89-91]. The Superoxide dismutase-1 (SOD1) transgenic mice remain the most commonly used model for preclinical pharmacological trials [92-96]. Whole-genome expression analysis has been performed in some of these animal models, and deregulated genes or pathways are available in genomic database for further studies [97, 98]. The comparison of published gene expression data from human SALS and mouse ALS models can help researchers prioritize drug candidates for validation processes. The selection of common genomic changes between human and different ALS models is therefore feasible, and represents a possible strategy to mitigate the currently high attrition rate of pharmacological compounds for ALS.
Fig. (8).
Drug target validation in mouse models of ALS. (A) Comparison between whole genome expression profiles of human SALS and mouse ALS models can provide the identification of common genomic changes. (B) Some established animal models of ALS.
CONCLUSIONS
With the completion of genome sequencing in humans and model organisms, together with the advent of high-throughput technologies, the transcriptional cascades and networks deregulated in ALS are being elucidated providing new potential pharmacological targets [36]. The computational techniques associated with enormous datasets of information derived by high-throughput technologies, will allow to portrait the altered networks of biological molecules in ALS and provide important insights into drug targets capable of interfering with ALS pathogenesis [99]. In view of the gene- and protein-signaling networks associated to complex disorder such as ALS, we have to rethink our strategies for drug development, targeting ALS pathogenesis as a system rather than at the level of the single protein molecule.
ACKNOWLEDGEMENT
The authors gratefully acknowledge Paolo Cantaro and Ambrogio Mazzeo for their encouragement, support and inspiration.
CONFLICT OF INTEREST
None declared.
REFERENCES
- 1.Mitchell JD, Borasio GD. Amyotrophic lateral sclerosis. Lancet. 2007;369(9578):2031–2041. doi: 10.1016/S0140-6736(07)60944-1. [DOI] [PubMed] [Google Scholar]
- 2.Aranda B, Achuthan P, Alam-Faruque Y, Armean I, Bridge A, Derow C, Feuermann M, Ghanbarian AT, Kerrien S, Khadake J, Kerssemakers J, Leroy C, Menden M, Michaut M, Montecchi-Palazzi L, Neuhauser SN, Orchard S, Perreau V, Roechert B, van Eijk K, Hermjakob H. The IntAct molecular interaction database in 2010. Nucleic Acids Res. 2010;38(Database issue):D525–531. doi: 10.1093/nar/gkp878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beghi E, Logroscino G, Chio A, Hardiman O, Mitchell D, Swingler R, Traynor BJ. The epidemiology of ALS and the role of population-based registries. Biochim. Biophys. Acta. 2006;1762(11-12 ):1150–1157. doi: 10.1016/j.bbadis.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 4.Miller RG, Mitchell JD, Moore DH. Riluzole for amyotrophic lateral sclerosis (ALS)/motor neuron disease (MND) (CD001447).Cochrane Database Syst. Rev. 2012;3 doi: 10.1002/14651858.CD001447. [DOI] [PubMed] [Google Scholar]
- 5.Pasinelli P, Brown RH. Molecular biology of amyotrophic lateral sclerosis: insights from genetics. Nat. Rev. Neurosci. 2006;7(9):710–723. doi: 10.1038/nrn1971. [DOI] [PubMed] [Google Scholar]
- 6.Rosen DR. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;364(6435):362. doi: 10.1038/364362c0. [DOI] [PubMed] [Google Scholar]
- 7.Chen YZ, Bennett CL, Huynh HM, Blair IP, Puls I, Irobi J, Dierick I, Abel A, Kennerson ML, Rabin BA, Nicholson GA, Auer-Grumbach M, Wagner K, De Jonghe P, Griffin JW, Fischbeck KH, Timmerman V, Cornblath DR, Chance PF. DNA/RNA helicase gene mutations in a form of juvenile amyotrophic lateral sclerosis (ALS4) Am. J. Hum. Genet. 2004;74(6):1128–1135. doi: 10.1086/421054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nishimura AL, Mitne-Neto M, Silva HC, Richieri-Costa A, Middleton S, Cascio D, Kok F, Oliveira JR, Gillingwater T, Webb J, Skehel P, Zatz M. A mutation in the vesicle-trafficking protein VAPB causes late-onset spinal muscular atrophy and amyotrophic lateral sclerosis. Am. J. Hum. Genet. 2004;75(5 ):822–831. doi: 10.1086/425287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hadano S, Hand CK, Osuga H, Yanagisawa Y, Otomo A, Devon RS, Miyamoto N, Showguchi-Miyata J, Okada Y, Singaraja R, Figlewicz DA, Kwiatkowski T, Hosler BA, Sagie T, Skaug J, Nasir J, Brown RH, Jr., Scherer SW, Rouleau GA, Hayden MR, Ikeda JE. A gene encoding a putative GTPase regulator is mutated in familial amyotrophic lateral sclerosis 2. Nat. Genet. 2001;29(2 ):166–173. doi: 10.1038/ng1001-166. [DOI] [PubMed] [Google Scholar]
- 10.Yang Y, Hentati A, Deng HX, Dabbagh O, Sasaki T, Hirano M, Hung WY, Ouahchi K, Yan J, Azim AC, Cole N, Gascon G, Yagmour A, Ben-Hamida M, Pericak-Vance M, Hentati F, Siddique T. The gene encoding alsin, a protein with three guanine-nucleotide exchange factor domains, is mutated in a form of recessive amyotrophic lateral sclerosis. Nat. Genet. 2001;29(2 ):160–165. doi: 10.1038/ng1001-160. [DOI] [PubMed] [Google Scholar]
- 11.Orlacchio A, Babalini C, Borreca A, Patrono C, Massa R, Basaran S, Munhoz RP, Rogaeva EA, St George-Hyslop PH, Bernardi G, Kawarai T. SPATACSIN mutations cause autosomal recessive juvenile amyotrophic lateral sclerosis. Brain. 2010;133(Pt 2 ):591–598. doi: 10.1093/brain/awp325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Greenway MJ, Andersen PM, Russ C, Ennis S, Cashman S, Donaghy C, Patterson V, Swingler R, Kieran D, Prehn J, Morrison KE, Green A, Acharya KR, Brown RH, Jr., Hardiman O. ANG mutations segregate with familial and 'sporadic' amyotrophic lateral sclerosis. Nat. Genet. 2006;38(4 ):411–413. doi: 10.1038/ng1742. [DOI] [PubMed] [Google Scholar]
- 13.Chow CY, Zhang Y, Dowling JJ, Jin N, Adamska M, Shiga K, Szigeti K, Shy ME, Li J, Zhang X, Lupski JR, Weisman LS, Meisler MH. Mutation of FIG4 causes neurodegeneration in the pale tremor mouse and patients with CMT4J. Nature. 2007;448(7149 ):68–72. doi: 10.1038/nature05876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maruyama H, Morino H, Ito H, Izumi Y, Kato H, Watanabe Y, Kinoshita Y, Kamada M, Nodera H, Suzuki H, Komure O, Matsuura S, Kobatake K, Morimoto N, Abe K, Suzuki N, Aoki M, Kawata A, Hirai T, Kato T, Ogasawara K, Hirano A, Takumi T, Kusaka H, Hagiwara K, Kaji R, Kawakami H. Mutations of optineurin in amyotrophic lateral sclerosis. Nature. 2010;465(7295 ):223–226. doi: 10.1038/nature08971. [DOI] [PubMed] [Google Scholar]
- 15.Majoor-Krakauer D, Willems PJ, Hofman A. Genetic epidemiology of amyotrophic lateral sclerosis. Clin. Genet. 2003;63(2 ):83–101. doi: 10.1046/j.0009-9163.2002.00001.x. [DOI] [PubMed] [Google Scholar]
- 16.Kabashi E, Valdmanis PN, Dion P, Spiegelman D, McConkey BJ, Vande Velde C, Bouchard JP, Lacomblez L, Pochigaeva K, Salachas F, Pradat PF, Camu W, Meininger V, Dupre N, Rouleau GA. TARDBP mutations in individuals with sporadic and familial amyotrophic lateral sclerosis. Nat. Genet. 2008;40(5 ):572–574. doi: 10.1038/ng.132. [DOI] [PubMed] [Google Scholar]
- 17.Vance C, Rogelj B, Hortobagyi T, De Vos KJ, Nishimura AL, Sreedharan J, Hu X, Smith B, Ruddy D, Wright P, Ganesalingam J, Williams KL, Tripathi V, Al-Saraj S, Al-Chalabi A, Leigh PN, Blair IP, Nicholson G, de Belleroche J, Gallo JM, Miller CC, Shaw CE. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science. 2009;323(5918 ):1208–1211. doi: 10.1126/science.1165942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schymick JC, Scholz SW, Fung HC, Britton A, Arepalli S, Gibbs JR, Lombardo F, Matarin M, Kasperaviciute D, Hernandez DG, Crews C, Bruijn L, Rothstein J, Mora G, Restagno G, Chio A, Singleton A, Hardy J, Traynor BJ. Genome-wide genotyping in amyotrophic lateral sclerosis and neurologically normal controls: first stage analysis and public release of data. Lancet Neurol. 2007;6(4 ):322–328. doi: 10.1016/S1474-4422(07)70037-6. [DOI] [PubMed] [Google Scholar]
- 19.van Es MA, Veldink JH, Saris CG, Blauw HM, van Vught PW, Birve A, Lemmens R, Schelhaas HJ, Groen EJ, Huisman MH, van der Kooi AJ, de Visser M, Dahlberg C, Estrada K, Rivadeneira F, Hofman A, Zwarts MJ, van Doormaal PT, Rujescu D, Strengman E, Giegling I, Muglia P, Tomik B, Slowik A, Uitterlinden AG, Hendrich C, Waibel S, Meyer T, Ludolph AC, Glass JD, Purcell S, Cichon S, Nothen MM, Wichmann HE, Schreiber S, Vermeulen SH, Kiemeney LA, Wokke JH, Cronin S, McLaughlin RL, Hardiman O, Fumoto K, Pasterkamp RJ, Meininger V, Melki J, Leigh PN, Shaw CE, Landers JE, Al-Chalabi A, Brown RH, Jr, Robberecht W, Andersen PM, Ophoff RA, van den Berg LH. Genome-wide association study identifies 19p13.(UNC13A) and 9p21.2 as susceptibility loci for sporadic amyotrophic lateral sclerosis. Nat. Genet. 2009;41(10 ):1083–1087. doi: 10.1038/ng.442. [DOI] [PubMed] [Google Scholar]
- 20.Laaksovirta H, Peuralinna T, Schymick JC, Scholz SW, Lai SL, Myllykangas L, Sulkava R, Jansson L, Hernandez DG, Gibbs JR, Nalls MA, Heckerman D, Tienari PJ, Traynor BJ. Chromosome 9p21 in amyotrophic lateral sclerosis in Finland: a genome-wide association study. Lancet Neurol. 2010;9(10 ):978–985. doi: 10.1016/S1474-4422(10)70184-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shatunov A, Mok K, Newhouse S, Weale ME, Smith B, Vance C, Johnson L, Veldink JH, van Es MA, van den Berg LH, Robberecht W, Van Damme P, Hardiman O, Farmer AE, Lewis CM, Butler AW, Abel O, Andersen PM, Fogh I, Silani V, Chio A, Traynor BJ, Melki J, Meininger V, Landers JE, McGuffin P, Glass JD, Pall H, Leigh PN, Hardy J, Brown RH, Jr., Powell JF, Orrell RW, Morrison KE, Shaw PJ, Shaw CE, Al-Chalabi A. Chromosome 9p21 in sporadic amyotrophic lateral sclerosis in the UK and seven other countries: a genome-wide association study. Lancet Neurol. 2010;9(10 ):986–994. doi: 10.1016/S1474-4422(10)70197-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van Deerlin VM, Sleiman PM, Martinez-Lage M, Chen-Plotkin A, Wang LS, Graff-Radford NR, Dickson DW, Rademakers R, Boeve BF, Grossman M, Arnold SE, Mann DM, Pickering-Brown SM, Seelaar H, Heutink P, van Swieten JC, Murrell JR, Ghetti B, Spina S, Grafman J, Hodges J, Spillantini MG, Gilman S, Lieberman AP, Kaye JA, Woltjer RL, Bigio EH, Mesulam M, Al-Sarraj S, Troakes C, Rosenberg RN, White CL 3rd, Ferrer I, Llado A, Neumann M, Kretzschmar HA, Hulette CM, Welsh-Bohmer KA, Miller BL, Alzualde A, Lopez de Munain A, McKee AC, Gearing M, Levey AI, Lah JJ, Hardy J, Rohrer JD, Lashley T, Mackenzie IR, Feldman HH, Hamilton RL, Dekosky ST, van der Zee J, Kumar-Singh S, Van Broeckhoven C, Mayeux R, Vonsattel JP, Troncoso JC, Kril JJ, Kwok JB, Halliday GM, Bird TD, Ince PG, Shaw PJ, Cairns NJ, Morris JC, McLean CA, DeCarli C, Ellis WG, Freeman SH, Frosch MP, Growdon JH, Perl DP, Sano M, Bennett DA, Schneider JA, Beach TG, Reiman EM, Woodruff BK, Cummings J, Vinters HV, Miller CA, Chui HC, Alafuzoff I, Hartikainen P, Seilhean D, Galasko D, Masliah E, Cotman CW, Tunon MT, Martinez MC, Munoz DG, Carroll SL, Marson D, Riederer PF, Bogdanovic N, Schellenberg GD, Hakonarson H, Trojanowski JQ, Lee VM. Common variants at 7p21 are associated with frontotemporal lobar degeneration with TDP-43 inclusions. Nat. Genet. 2010;42(3 ):234–239. doi: 10.1038/ng.536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.DeJesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, Nicholson AM, Finch NA, Flynn H, Adamson J, Kouri N, Wojtas A, Sengdy P, Hsiung GY, Karydas A, Seeley WW, Josephs KA, Coppola G, Geschwind DH, Wszolek ZK, Feldman H, Knopman DS, Petersen RC, Miller BL, Dickson DW, Boylan KB, Graff-Radford NR, Rademakers R. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72(2 ):245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Renton AE, Majounie E, Waite A, Simon-Sanchez J, Rollinson S, Gibbs JR, Schymick JC, Laaksovirta H, van Swieten JC, Myllykangas L, Kalimo H, Paetau A, Abramzon Y, Remes AM, Kaganovich A, Scholz SW, Duckworth J, Ding J, Harmer DW, Hernandez DG, Johnson JO, Mok K, Ryten M, Trabzuni D, Guerreiro RJ, Orrell RW, Neal J, Murray A, Pearson J, Jansen IE, Sondervan D, Seelaar H, Blake D, Young K, Halliwell N, Callister JB, Toulson G, Richardson A, Gerhard A, Snowden J, Mann D, Neary D, Nalls MA, Peuralinna T, Jansson L, Isoviita VM, Kaivorinne AL, Holtta-Vuori M, Ikonen E, Sulkava R, Benatar M, Wuu J, Chio A, Restagno G, Borghero G, Sabatelli M, Heckerman D, Rogaeva E, Zinman L, Rothstein JD, Sendtner M, Drepper C, Eichler EE, Alkan C, Abdullaev Z, Pack SD, Dutra A, Pak E, Hardy J, Singleton A, Williams NM, Heutink P, Pickering-Brown S, Morris HR, Tienari PJ, Traynor BJ. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72(2 ):257–268. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ferraiuolo L, Heath PR, Holden H, Kasher P, Kirby J, Shaw PJ. Microarray analysis of the cellular pathways involved in the adaptation to and progression of motor neuron injury in the SOD1 G93A mouse model of familial ALS. J. Neurosci. 2007;27(34 ):9201–9219. doi: 10.1523/JNEUROSCI.1470-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fukada Y, Yasui K, Kitayama M, Doi K, Nakano T, Watanabe Y, Nakashima K. Gene expression analysis of the murine model of amyotrophic lateral sclerosis: studies of the Leu126delTT mutation in SOD1. Brain Res. 2007;1160:1–10. doi: 10.1016/j.brainres.2007.05.044. [DOI] [PubMed] [Google Scholar]
- 27.Hensley K, Floyd RA, Gordon B, Mou S, Pye QN, Stewart C, West M, Williamson K. Temporal patterns of cytokine and apoptosis-related gene expression in spinal cords of the G93A-SOD1 mouse model of amyotrophic lateral sclerosis. J. Neurochem. 2002;82(2 ):365–374. doi: 10.1046/j.1471-4159.2002.00968.x. [DOI] [PubMed] [Google Scholar]
- 28.Lobsiger CS, Boillee S, Cleveland DW. Toxicity from different SOD1 mutants dysregulates the complement system and the neuronal regenerative response in ALS motor neurons. Proc. Natl. Acad. Sci. USA. 2007;104(18 ):7319–7326. doi: 10.1073/pnas.0702230104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yoshihara T, Ishigaki S, Yamamoto M, Liang Y, Niwa J, Takeuchi H, Doyu M, Sobue G. Differential expression of inflammation- and apoptosis-related genes in spinal cords of a mutant SOD1 transgenic mouse model of familial amyotrophic lateral sclerosis. J. Neurochem. 2002;80(1 ):158–167. doi: 10.1046/j.0022-3042.2001.00683.x. [DOI] [PubMed] [Google Scholar]
- 30.Dangond F, Hwang D, Camelo S, Pasinelli P, Frosch MP, Stephanopoulos G, Brown RH, Jr, Gullans SR. Molecular signature of late-stage human ALS revealed by expression profiling of postmortem spinal cord gray matter. Physiol. Genomics. 2004;16(2 ):229–239. doi: 10.1152/physiolgenomics.00087.2001. [DOI] [PubMed] [Google Scholar]
- 31.Ishigaki S, Niwa J, Ando Y, Yoshihara T, Sawada K, Doyu M, Yamamoto M, Kato K, Yotsumoto Y, Sobue G. Differentially expressed genes in sporadic amyotrophic lateral sclerosis spinal cords--screening by molecular indexing and subsequent cDNA microarray analysis. FEBS Lett. 2002;531(2 ):354–358. doi: 10.1016/s0014-5793(02)03546-9. [DOI] [PubMed] [Google Scholar]
- 32.Malaspina A, Kaushik N, de Belleroche J. Differential expression of 14 genes in amyotrophic lateral sclerosis spinal cord detected using gridded cDNA arrays. J. Neurochem. 2001;77(1 ):132–145. doi: 10.1046/j.1471-4159.2001.t01-1-00231.x. [DOI] [PubMed] [Google Scholar]
- 33.Offen D, Barhum Y, Melamed E, Embacher N, Schindler C, Ransmayr G. Spinal cord mRNA profile in patients with ALS: comparison with transgenic mice expressing the human SOD-1 mutant. J. Mol. Neurosci. 2009;38(2 ):85–93. doi: 10.1007/s12031-007-9004-z. [DOI] [PubMed] [Google Scholar]
- 34.Jiang YM, Yamamoto M, Kobayashi Y, Yoshihara T, Liang Y, Terao S, Takeuchi H, Ishigaki S, Katsuno M, Adachi H, Niwa J, Tanaka F, Doyu M, Yoshida M, Hashizume Y, Sobue G. Gene expression profile of spinal motor neurons in sporadic amyotrophic lateral sclerosis. Ann.Neurol. 2005;57(2 ):236–251. doi: 10.1002/ana.20379. [DOI] [PubMed] [Google Scholar]
- 35.Wang XS, Simmons Z, Liu W, Boyer PJ, Connor JR. Differential expression of genes in amyotrophic lateral sclerosis revealed by profiling the post mortem cortex. Amyotroph. Lateral. Scler. 2006;7(4 ):201–210. doi: 10.1080/17482960600947689. [DOI] [PubMed] [Google Scholar]
- 36.Lederer CW, Torrisi A, Pantelidou M, Santama N, Cavallaro S. Pathways and genes differentially expressed in the motor cortex of patients with sporadic amyotrophic lateral sclerosis. BMC Genomics. 2007;8(1 ):26. doi: 10.1186/1471-2164-8-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kudo LC, Parfenova L, Vi N, Lau K, Pomakian J, Valdmanis P, Rouleau GA, Vinters HV, Wiedau-Pazos M, Karsten SL. Integrative gene-tissue microarray-based approach for identification of human disease biomarkers: application to amyotrophic lateral sclerosis. Hum. Mol. Genet. 2010;19(16 ):3233–3253. doi: 10.1093/hmg/ddq232. [DOI] [PubMed] [Google Scholar]
- 38.Petri S, Krampfl K, Hashemi F, Grothe C, Hori A, Dengler R, Bufler J. Distribution of GABAA receptor mRNA in the motor cortex of ALS patients. J. Neuropathol. Exp. Neurol. 2003;62(10 ):1041–1051. doi: 10.1093/jnen/62.10.1041. [DOI] [PubMed] [Google Scholar]
- 39.de Carvalho Aguiar P, Sweadner KJ, Penniston JT, Zaremba J, Liu L, Caton M, Linazasoro G, Borg M, Tijssen MA, Bressman SB, Dobyns WB, Brashear A, Ozelius LJ. Mutations in the Na+/K+ -ATPase alpha3 gene ATP1A3 are associated with rapid-onset dystonia parkinsonism. Neuron. 2004;43(2 ):169–175. doi: 10.1016/j.neuron.2004.06.028. [DOI] [PubMed] [Google Scholar]
- 40.Ellis DZ, Rabe J, Sweadner KJ. Global loss of Na,K-ATPase and its nitric oxide-mediated regulation in a transgenic mouse model of amyotrophic lateral sclerosis. J. Neurosci. 2003;23(1 ):43–51. doi: 10.1523/JNEUROSCI.23-01-00043.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gonzalez Deniselle MC, Lopez-Costa JJ, Saavedra JP, Pietranera L, Gonzalez SL, Garay L, Guennoun R, Schumacher M, De Nicola AF. Progesterone neuroprotection in the Wobbler mouse, a genetic model of spinal cord motor neuron disease. Neurobiol. Dis. 2002;11(3 ):457–468. doi: 10.1006/nbdi.2002.0564. [DOI] [PubMed] [Google Scholar]
- 42.Wang JW, Humphreys JM, Phillips JP, Hilliker AJ, Wu CF. novel leg-shaking Drosophila mutant defective in a voltage-gated K(+)current and hypersensitive to reactive oxygen species. J. Neurosci. 2000;20(16 ):5958–5964. doi: 10.1523/JNEUROSCI.20-16-05958.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Blaesse P, Airaksinen MS, Rivera C, Kaila K. ation-chloride cotransporters and neuronal function. Neuron. 2009;61(6 ):820–838. doi: 10.1016/j.neuron.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 44.Rivera C, Voipio J, Payne JA, Ruusuvuori E, Lahtinen H, Lamsa K, Pirvola U, Saarma M, Kaila K. The K+/Cl- cotransporter KCC2 renders GABA hyperpolarizing during neuronal maturation. Nature. 1999;397(6716 ):251–255. doi: 10.1038/16697. [DOI] [PubMed] [Google Scholar]
- 45.Toyoda H, Ohno K, Yamada J, Ikeda M, Okabe A, Sato K, Hashimoto K, Fukuda A. Induction of NMDA and GABAA receptor-mediated Ca2+ oscillations with KCC2 mRNA downregulation in injured facial motoneurons. J. Neurophysiol. 2003;89(3 ):1353–1362. doi: 10.1152/jn.00721.2002. [DOI] [PubMed] [Google Scholar]
- 46.Nabekura J, Ueno T, Okabe A, Furuta A, Iwaki T, Shimizu-Okabe C, Fukuda A, Akaike N. Reduction of KCC2 expression and GABAA receptor-mediated excitation after in vivo axonal injury. J. Neurosci. 2002;22(11 ):4412–4417. doi: 10.1523/JNEUROSCI.22-11-04412.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hubner CA, Stein V, Hermans-Borgmeyer I, Meyer T, Ballanyi K, Jentsch TJ. Disruption of KCC2 reveals an essential role of K-Cl cotransport already in early synaptic inhibition. Neuron. 2001;30(2 ):515–524. doi: 10.1016/s0896-6273(01)00297-5. [DOI] [PubMed] [Google Scholar]
- 48.Roth FP, Lipshitz HD, Andrews BJ. Q&A: epistasis. J. Biol. 2009;8(4 ):35. doi: 10.1186/jbiol144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Harris MA, Clark J, Ireland A, Lomax J, Ashburner M, Foulger R, Eilbeck K, Lewis S, Marshall B, Mungall C, Richter J, Rubin GM, Blake JA, Bult C, Dolan M, Drabkin H, Eppig JT, Hill DP, Ni L, Ringwald M, Balakrishnan R, Cherry JM, Christie KR, Costanzo MC, Dwight SS, Engel S, Fisk DG, Hirschman JE, Hong EL, Nash RS, Sethuraman A, Theesfeld CL, Botstein D, Dolinski K, Feierbach B, Berardini T, Mundodi S, Rhee SY, Apweiler R, Barrell D, Camon E, Dimmer E, Lee V, Chisholm R, Gaudet P, Kibbe W, Kishore R, Schwarz EM, Sternberg P, Gwinn M, Hannick L, Wortman J, Berriman M, Wood V, de la Cruz N, Tonellato P, Jaiswal P, Seigfried T, White R. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 2004;32(Database issue ):D258–261. doi: 10.1093/nar/gkh036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kanehisa M. The KEGG database.discussion 101-103 119-128: 244-152. Novartis Found. Symp. 2002;247:91–101. [PubMed] [Google Scholar]
- 51.Dahlquist KD, Salomonis N, Vranizan K, Lawlor SC, Conklin BR. GenMAPP, a new tool for viewing and analyzing microarray data on biological pathways. Nat. Genet. 2002;31(1 ):19–20. doi: 10.1038/ng0502-19. [DOI] [PubMed] [Google Scholar]
- 52.Shaw PJ. Molecular and cellular pathways of neurodegeneration in motor neurone disease. J Neurol. Neurosurg. Psychiatry. 2005;76(8 ):1046–1057. doi: 10.1136/jnnp.2004.048652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhao C, Takita J, Tanaka Y, Setou M, Nakagawa T, Takeda S, Yang HW, Terada S, Nakata T, Takei Y, Saito M, Tsuji S, Hayashi Y, Hirokawa N. Charcot-Marie-Tooth disease type 2A caused by mutation in a microtubule motor KIF1Bbeta. Cell. 2001;105(5 ):587–597. doi: 10.1016/s0092-8674(01)00363-4. [DOI] [PubMed] [Google Scholar]
- 54.Reid E, Kloos M, Ashley-Koch A, Hughes L, Bevan S, Svenson IK, Graham FL, Gaskell PC, Dearlove A, Pericak-Vance MA, Rubinsztein DC, Marchuk DA. A kinesin heavy chain (KIF5A) mutation in hereditary spastic paraplegia (SPG10) Am. J. Hum. Genet. 2002;71(5 ):1189–1194. doi: 10.1086/344210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hurd DD, Saxton WM. Kinesin mutations cause motor neuron disease phenotypes by disrupting fast axonal transport in Drosophila. Genetics. 1996;144(3 ):1075–1085. doi: 10.1093/genetics/144.3.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.LaMonte BH, Wallace KE, Holloway BA, Shelly SS, Ascano J, Tokito M, Van Winkle T, Howland DS, Holzbaur EL. Disruption of dynein/dynactin inhibits axonal transport in motor neurons causing late-onset progressive degeneration. Neuron. 2002;34(5 ):715–727. doi: 10.1016/s0896-6273(02)00696-7. [DOI] [PubMed] [Google Scholar]
- 57.Williamson TL, Bruijn LI, Zhu Q, Anderson KL, Anderson SD, Julien JP, Cleveland DW. Absence of neurofilaments reduces the selective vulnerability of motor neurons and slows disease caused by a familial amyotrophic lateral sclerosis-linked superoxide dismutase 1 mutant. Proc. Natl. Acad. Sci. U S A. 1998;95(16 ):9631–9636. doi: 10.1073/pnas.95.16.9631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Couillard-Despres S, Zhu Q, Wong PC, Price DL, Cleveland DW, Julien JP. Protective effect of neurofilament heavy gene overexpression in motor neuron disease induced by mutant superoxide dismutase. Proc. Natl. Acad. Sci. U S A. 1998;95(16 ):9626–9630. doi: 10.1073/pnas.95.16.9626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bruijn LI, Beal MF, Becher MW, Schulz JB, Wong PC, Price DL, Cleveland DW. Elevated free nitrotyrosine levels, but not protein-bound nitrotyrosine or hydroxyl radicals, throughout amyotrophic lateral sclerosis (ALS)-like disease implicate tyrosine nitration as an aberrant in vivo property of one familial ALS-linked superoxide dismutase 1 mutant. Proc. Natl. Acad. Sci. U S A. 1997;94(14 ):7606–7611. doi: 10.1073/pnas.94.14.7606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Julien JP, Beaulieu JM. Cytoskeletal abnormalities in amyotrophic lateral sclerosis: beneficial or detrimental effects? J. Neurol. Sci. 2000;180(1-2 ):7–14. doi: 10.1016/s0022-510x(00)00422-6. [DOI] [PubMed] [Google Scholar]
- 61.Williamson TL, Cleveland DW. Slowing of axonal transport is a very early event in the toxicity of ALS-linked SOD1 mutants to motor neurons. Nat. Neurosci. 1999;2(1 ):50–56. doi: 10.1038/4553. [DOI] [PubMed] [Google Scholar]
- 62.Costanzo M, Baryshnikova A, Bellay J, Kim Y, Spear ED, Sevier CS, Ding H, Koh JL, Toufighi K, Mostafavi S, Prinz J, St Onge RP, VanderSluis B, Makhnevych T, Vizeacoumar FJ, Alizadeh S, Bahr S, Brost RL, Chen Y, Cokol M, Deshpande R, Li Z, Lin ZY, Liang W, Marback M, Paw J, San Luis BJ, Shuteriqi E, Tong AH, van Dyk N, Wallace IM, Whitney JA, Weirauch MT, Zhong G, Zhu H, Houry WA, Brudno M, Ragibizadeh S, Papp B, Pal C, Roth FP, Giaever G, Nislow C, Troyanskaya OG, Bussey H, Bader GD, Gingras AC, Morris QD, Kim PM, Kaiser CA, Myers CL, Andrews BJ, Boone C. The genetic landscape of a cell. Science. 2010;327(5964 ):425–431. doi: 10.1126/science.1180823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sauer U, Heinemann M, Zamboni N. Genetics. Getting closer to the whole picture. Science. 2007;316(5824 ):550–551. doi: 10.1126/science.1142502. [DOI] [PubMed] [Google Scholar]
- 64.Seeley WW, Crawford RK, Zhou J, Miller BL, Greicius MD. Neurodegenerative diseases target large-scale human brain networks. Neuron. 2009;62(1 ):42–52. doi: 10.1016/j.neuron.2009.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Limviphuvadh V, Tanaka S, Goto S, Ueda K, Kanehisa M. The commonality of protein interaction networks determined in neurodegenerative disorders (NDDs) Bioinformatics. 2007;23(16 ):2129–2138. doi: 10.1093/bioinformatics/btm307. [DOI] [PubMed] [Google Scholar]
- 66.Morphy R, Kay C, Rankovic Z. From magic bullets to designed multiple ligands. Drug Discov. Today. 2004;9(15 ):641–651. doi: 10.1016/S1359-6446(04)03163-0. [DOI] [PubMed] [Google Scholar]
- 67.Stark C, Breitkreutz BJ, Chatr-Aryamontri A, Boucher L, Oughtred R, Livstone MS, Nixon J, Van Auken K, Wang X, Shi X, Reguly T, Rust JM, Winter A, Dolinski K, Tyers M. The BioGRID Interaction Database: 2011 update. Nucleic Acids Res. 2011;39(Database issue ):D698–704. doi: 10.1093/nar/gkq1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ceol A, Chatr Aryamontri A, Licata L, Peluso D, Briganti L, Perfetto L, Castagnoli L, Cesareni G. MINT, the molecular interaction database: 2009 update. Nucleic Acids Res. 2010;38(Database issue):D532–539. doi: 10.1093/nar/gkp983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ekins S, Bugrim A, Brovold L, Kirillov E, Nikolsky Y, Rakhmatulin E, Sorokina S, Ryabov A, Serebryiskaya T, Melnikov A, Metz J, Nikolskaya T. Algorithms for network analysis in systems-ADME/Tox using the MetaCore and MetaDrug platforms. Xenobiotica. 2006;36(10-11 ):877–901. doi: 10.1080/00498250600861660. [DOI] [PubMed] [Google Scholar]
- 70.Goel R, Muthusamy B, Pandey A, Prasad TS. Human protein reference database and human proteinpedia as discovery resources for molecular biotechnology. Mol. Biotechnol. 2011;48(1 ):87–95. doi: 10.1007/s12033-010-9336-8. [DOI] [PubMed] [Google Scholar]
- 71.Salwinski L, Miller CS, Smith AJ, Pettit FK, Bowie JU, Eisenberg D. The Database of Interacting Proteins: 2004 update. Nucleic Acids Res. 2004;32(Database issue):D449–451. doi: 10.1093/nar/gkh086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Palmiter RD. The elusive function of metallothioneins. Proc. Natl. Acad. Sci. U S A. 1998;95(15):8428–8430. doi: 10.1073/pnas.95.15.8428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kelly EJ, Quaife CJ, Froelick GJ, Palmiter RD. Metallothionein I and II protect against zinc deficiency and zinc toxicity in mice. J. Nutr. 1996;126(7 ):1782–1790. doi: 10.1093/jn/126.7.1782. [DOI] [PubMed] [Google Scholar]
- 74.Kim J, Kim TY, Hwang JJ, Lee JY, Shin JH, Gwag BJ, Koh JY. Accumulation of labile zinc in neurons and astrocytes in the spinal cords of G93A SOD-1 transgenic mice. Neurobiol. Dis. 2009;34(2 ):221–229. doi: 10.1016/j.nbd.2009.01.004. [DOI] [PubMed] [Google Scholar]
- 75.Liu Y, Liu J, Iszard MB, Andrews GK, Palmiter RD, Klaassen CD. Transgenic mice that overexpress metallothionein-I are protected from cadmium lethality and hepatotoxicity. Toxicol. Appl. Pharmacol. 1995;135(2 ):222–228. doi: 10.1006/taap.1995.1227. [DOI] [PubMed] [Google Scholar]
- 76.Taylor DM, Minotti S, Agar JN, Durham HD. Overexpression of metallothionein protects cultured motor neurons against oxidative stress, but not mutant Cu/Zn-superoxide dismutase toxicity. Neurotoxicology. 2004;25(5 ):779–792. doi: 10.1016/j.neuro.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 77.Chung RS, Penkowa M, Dittmann J, King CE, Bartlett C, Asmussen JW, Hidalgo J, Carrasco J, Leung YK, Walker AK, Fung SJ, Dunlop SA, Fitzgerald M, Beazley LD, Chuah MI, Vickers JC, West AK. Redefining the role of metallothionein within the injured brain: extracellular metallothioneins play an important role in the astrocyte-neuron response to injury. J. Biol. Chem. 2008;283(22 ):15349–15358. doi: 10.1074/jbc.M708446200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fauman EB, Rai BK, Huang ES. Structure-based druggability assessment--identifying suitable targets for small molecule therapeutics. Curr. Opin. Chem. Biol. 2011;15(4 ):463–468. doi: 10.1016/j.cbpa.2011.05.020. [DOI] [PubMed] [Google Scholar]
- 79.Hopkins AL, Groom CR. The druggable genome. Nat. Rev. Drug Discov. 2002;1(9):727–730. doi: 10.1038/nrd892. [DOI] [PubMed] [Google Scholar]
- 80.Yildirim MA, Goh KI, Cusick ME, Barabasi AL, Vidal M. Drug-target network. Nat. Biotechnol. 2007;25(10):1119–1126. doi: 10.1038/nbt1338. [DOI] [PubMed] [Google Scholar]
- 81.Knox C, Law V, Jewison T, Liu P, Ly S, Frolkis A, Pon A, Banco K, Mak C, Neveu V, Djoumbou Y, Eisner R, Guo AC, Wishart DS. DrugBank 3.: a comprehensive resource for 'omics' research on drugs. Nucleic Acids Res. 2011;39(Database issue):D1035–1041. doi: 10.1093/nar/gkq1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhu F, Han B, Kumar P, Liu X, Ma X, Wei X, Huang L, Guo Y, Han L, Zheng C, Chen Y. Update of TTD: Therapeutic Target Database. Nucleic Acids Res. 2010;38(Database issue):D787–791. doi: 10.1093/nar/gkp1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Thorn CF, Klein TE, Altman RB. Pharmacogenomics and bioinformatics: PharmGKB. Pharmacogenomics. 2010;11(4 ):501–505. doi: 10.2217/pgs.10.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kuhn M, Szklarczyk D, Franceschini A, Campillos M, von Mering C, Jensen LJ, Beyer A, Bork P. STITCH 2: an interaction network database for small molecules and proteins. Nucleic Acids Res. 2010;38(Database issue):D552–556. doi: 10.1093/nar/gkp937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gunther S, Kuhn M, Dunkel M, Campillos M, Senger C, Petsalaki E, Ahmed J, Urdiales EG, Gewiess A, Jensen LJ, Schneider R, Skoblo R, Russell RB, Bourne PE, Bork P, Preissner R. SuperTarget and Matador: resources for exploring drug-target relationships. Nucleic Acids Res. 2008;36(Database issue):D919–922. doi: 10.1093/nar/gkm862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Suzuki S, Masui Y, Ohnuki M, Miyakoda G, Mori T, Nakajima K, Sato M. Induction of metallothionein synthesis by cilostazol in mice and in human cultured neuronal cell lines. Biol. Pharm. Bull. 2007;30(4 ):791–794. doi: 10.1248/bpb.30.791. [DOI] [PubMed] [Google Scholar]
- 87.Wakida K, Morimoto N, Shimazawa M, Hozumi I, Nagase H, Inuzuka T, Hara H. Cilostazol reduces ischemic brain damage partly by inducing metallothionein-1 and -2. Brain Res. 2006;1116(1 ):187–193. doi: 10.1016/j.brainres.2006.07.125. [DOI] [PubMed] [Google Scholar]
- 88.Kola I, Landis J. Can the pharmaceutical industry reduce attrition rates? Nat. Rev. Drug Discov. 2004;3(8 ):711–715. doi: 10.1038/nrd1470. [DOI] [PubMed] [Google Scholar]
- 89.Zhou H, Huang C, Chen H, Wang D, Landel CP, Xia PY, Bowser R, Liu YJ, Xia XG. Transgenic rat model of neurodegeneration caused by mutation in the TDP gene. (e1000887).PLoS Genet. 2010;6(3 ) doi: 10.1371/journal.pgen.1000887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Xu YF, Gendron TF, Zhang YJ, Lin WL, D'Alton S, Sheng H, Casey MC, Tong J, Knight J, Yu X, Rademakers R, Boylan K, Hutton M, McGowan E, Dickson DW, Lewis J, Petrucelli L. Wild-type human TDP-43 expression causes TDP-43 phosphorylation, mitochondrial aggregation, motor deficits, and early mortality in transgenic mice. J. Neurosci. 2010;30(32 ):10851–10859. doi: 10.1523/JNEUROSCI.1630-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Huang C, Zhou H, Tong J, Chen H, Liu YJ, Wang D, Wei X, Xia XG. FUS transgenic rats develop the phenotypes of amyotrophic lateral sclerosis and frontotemporal lobar degeneration. (e1002011).PLoS Genet. 2011;7(3 ) doi: 10.1371/journal.pgen.1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Swarup V, Julien JP. ALS pathogenesis: Recent insights from genetics and mouse models. Prog Neuropsychopharmacol. Biol. Psychiatry. 2010. [DOI] [PubMed]
- 93.Pioro EP, Mitsumoto H. Animal models of ALS. Clin. Neurosci. 1995;3(6 ):375–385. [PubMed] [Google Scholar]
- 94.Doble A, Kennel P. Animal models of amyotrophic lateral sclerosis. Amyotroph. Lateral Scler. Other Motor Neuron Disord. 2000;1(5 ):301–312. doi: 10.1080/146608200300079545. [DOI] [PubMed] [Google Scholar]
- 95.Cai H, Shim H, Lai C, Xie C, Lin X, Yang WJ, Chandran J. ALS2/alsin knockout mice and motor neuron diseases. Neurodegener. Dis. 2008;5(6 ):359–366. doi: 10.1159/000151295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wegorzewska I, Bell S, Cairns NJ, Miller TM, Baloh RH. TDP-43 mutant transgenic mice develop features of ALS and frontotemporal lobar degeneration. Proc. Natl. Acad. Sci. U S A. 2009;106(44 ):18809–18814. doi: 10.1073/pnas.0908767106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Barrett T, Edgar R. Gene expression omnibus: microarray data storage, submission, retrieval, and analysis. Methods Enzymol. 2006;411:352–369. doi: 10.1016/S0076-6879(06)11019-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Parkinson H, Sarkans U, Kolesnikov N, Abeygunawardena N, Burdett T, Dylag M, Emam I, Farne A, Hastings E, Holloway E, Kurbatova N, Lukk M, Malone J, Mani R, Pilicheva E, Rustici G, Sharma A, Williams E, Adamusiak T, Brandizi M, Sklyar N, Brazma A. ArrayExpress update--an archive of microarray and high-throughput sequencing-based functional genomics experiments. Nucleic Acids Res. 2011;39(Database issue):D1002–1004. doi: 10.1093/nar/gkq1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hopkins AL. Network pharmacology: the next paradigm in drug discovery. Nat. Chem. Biol. 2008;4(11 ):682–690. doi: 10.1038/nchembio.118. [DOI] [PubMed] [Google Scholar]