Abstract
Accurate and reliable quantitative proteomics in cell culture has been considerably facilitated by the introduction of the stable isotope labeling by amino acids in cell culture (SILAC), combined with high resolution mass spectrometry. There are however several major sources of quantification errors that commonly occur with SILAC techniques, i.e. incomplete incorporation of isotopic amino acids, arginine-to-proline conversion, and experimental errors in final sample mixing. Dataset normalization is a widely adopted solution to such errors, however this may not completely prevent introducing incorrect expression ratios. Here we demonstrate that a label-swap replication of SILAC experiments was able to effectively correct experimental errors by averaging ratios measured in individual replicates using quantitative proteomics and phosphoproteomics of ligand treatment of neural cell cultures. Furthermore, this strategy was successfully applied to a SILAC triplet experiment, which presents a much more complicated experimental matrix, affected by both incomplete labeling and arginine-to-proline conversion. Based on our results, we suggest that SILAC experiments should be designed to incorporate label-swap replications for enhanced reliability in expression ratios.
Keywords: SILAC, incomplete isotope labeling, arginine-to-proline conversion, label-swap replication, receptor
1. Introduction
Quantification is one of the most important issues in MS-based proteomics and has evolved tremendously in the last decade through the introduction of several isotope labeling methods, such as ICAT, iTRAQ, TMT (tandem mass tags) and SILAC (stable isotope labeling by amino acids in cell culture) [1–4]. In SILAC, stable isotope-labeled amino acids are incorporated into cellular proteins through endogenous protein synthesis pathways, allowing accurate quantification of all native proteins without any subsequent chemical modification. When it was first introduced, leucine (leu-D3) was employed because it is an essential amino acid [4]. Since then, isotope-labeled arginine and lysine have been successfully introduced and now almost all SILAC applications employ these two amino acids, as trypsin-digested peptides contain at least one arginine or lysine therefore rendering all peptides eligible for quantification [5, 6].
Despite the successful introduction of SILAC, there are however several major sources of quantification error with this technique, such as incomplete incorporation of isotopic amino acids, arginine-to-proline conversion, and experimental errors in the final sample mixing. Arginine in culture media is taken up by the recipient cells and can be converted to proline through the catalytic action of arginase. This can therefore result in the generation of proteins with proline residues that are also isotope-labeled with 13C and/or 15N from isotopic arginine [6, 7]. This conversion process can lead to decreased ion intensities of the ‘heavy’ (i.e. isotopically labeled) peptides and resultant reduced ‘heavy’/‘light (non-labeled)’ (H/L) ratios. Cells possessing a robust arginase activity are likely to exhibit, after SILAC labeling, the undesired incorporation of isotopic labels into proline, glutamate, glutamine and lysine, therefore imposing complicated quantification errors [8].
At present, there are four approaches that have been proposed to solve or attenuate such errors. The simplest mechanism may be through reduction in the arginine concentration in the culture media. This would effectively reduce the functional catalytic drive to the arginase pathway, however this amino acid reduction mechanism is likely to introduce new complications through perturbation of the balance of arginine-dependent amino acid synthesis in the cells. In contrast, altering the equilibrium from the other side of the arginine conversion process, i.e. the addition of proline to culture media, has been demonstrated to be effective in attenuating the rate of arginine-to-proline conversion in some cell lines [9]. These methods are primarily based upon substrate or product inhibition of enzyme activity in metabolic/synthetic pathways. These methods however may not be universally applicable as some cell lines or organisms may possess an extremely robust rate of arginine-to-proline conversion. In yeast (Schizosaccharomyces pombe), a solution through genetic engineering has been demonstrated, in which a key enzyme in the arginine-to-proline conversion process was genetically engineered to be nonfunctional [8]. This strategy, while elegant in its execution and extremely stable after implementation, is mainly limited to organisms amenable to facile genetic manipulation. Another approach recently developed involves the introduction of an additional isotope-labeled arginine ([15N4]-arginine) in the light condition so that heavy proline would be formed at the same rate under both light and heavy conditions, thus providing an internal correction for arginine conversion [10]. However, this process may be much more complicated in SILAC triplet experiments using three conditions of light, medium ([D4]-lysine, [13C6]-arginine) and heavy ([13C615N2]-lysine, [13C615N4]-arginine) and cost additionally.
In addition to the analytical complications introduced by arginine-to-proline conversion, the incomplete incorporation of isotopic amino acids and experimental mixing errors present another commonly experienced source of MS quantification errors. For example, the introduction of SILAC labels into cells is often problematic for primary cells (due to longevity and replication time) and for other cultured clonal cells possessing a high basal arginase activity. In these circumstances it is often considered prudent to apply dataset normalization procedures to account for potential errors, yet this still does not completely prevent generation of incorrect expression ratios. In this study, we propose a simple strategy, SILAC label-swap replication, readily applicable to all cultured cell lines or organisms to attenuate the SILAC experimental errors. Such a strategy has been employed for quantitative proteomic studies but its effect on experimental errors by incomplete labeling and arginine-to-proline conversion remained to be fully understood [11–13]. We applied SILAC label-swap replication and examined ratio distribution of each replicate and their average and found that errors in each replicate could be efficiently compensated for through geometric averaging. Moreover, we have demonstrated that the ratio averaging of label-swap replicates can be successfully extended to even a highly complex SILAC triplet experiment, thereby indicating the effective flexibility of our procedure.
2. Materials and Methods
2.1. Cell culture and treatment
Human neuroblastoma BE(2)-M17 cells were maintained in DMEM media (Invitrogen) supplemented with 10% dialyzed fetal bovine serum and 1% penicillin-streptomycin (Invitrogen). Customized DMEM media, without arginine and lysine, were purchased from AthenaES. L-lysine-D4 (K4), L-lysine-U-13C6-15N2 (K8), L-arginine-U-13C6 (R6) and L-arginine-U-13C6-15N4 (R10) were purchased from Cambridge Isotope Laboratories. For experiments involving proline addition, L-proline (Sigma-Aldrich) was added to the DMEM media. For ligand treatment, cells were grown for 7 days (eight division cycles) in DMEM media containing light (L: K0R0), medium (M: K4R6) or heavy (H: K8R10) forms of arginine and lysine and then treated with 10 ng/mL EGF for 10 min, 10 μM pilocarpine for 5 min, or 10 μM muscarine (Sigma-Aldrich) for 1, 5, and 20 min.
2.2. Peptide sample preparation
After treatment, cells were washed three times with ice-cold PBS and scraped from dishes in the presence of a lysis buffer and completely disrupted using a Sonic Dismembrator (Model 100; Thermo Fisher Scientific). The lysis buffer was prepared with 8 M urea, 50 mM Tris pH 8.0, 75 mM NaCl, 1 mM NaF, 1 mM β-glycerophosphate, 1 mM sodium orthovanadate, 10 mM sodium pyrophosphate, 1 mM phenylmethanesulfonylfluoride and one tablet of protease inhibitors cocktail (complete mini, Roche Diagnostics) per 10 mL of lysis buffer. Resultant protein lysate concentrations were determined by BCA protein assay kit (Thermo Fisher Scientific). Equal amounts or designated ratio of proteins from light, medium or heavy conditions were mixed to prepare SILAC doublet or triplet mix. Proteins were reduced in 10 mM DTT at room temperature (RT) for 30 min and alkylated with 25 mM iodoacetamide at RT for 20 min in the dark. The reaction was quenched with an additional incubation in 15 mM DTT at RT for 15 min. Protein extracts were diluted in 50 mM Tris pH 8.0 to a final concentration of 1.5 M urea and were digested at 37°C using sequencing grade trypsin (Promega) at a 1:100 ratio (trypsin/protein, w/w). After overnight digestion, formic acid was added to a final concentration of 0.5% (v/v) and the peptide samples were desalted using Sep-Pak Vac C18 Cartridges. Cartridges were washed with ACN and equilibrated with 0.1% formic acid. After loading peptides, cartridges were washed with 0.1% formic acid. Peptides were then eluted with 70% ACN 0.1% formic acid, dried but not completely in SpeedVac (Thermo Fisher Scientific), and stored at −80°C until used.
2.3. Phosphopeptide enrichment
The preparation of TiO2 capillary columns was performed as described previously [14]. TiO2 particles (GL sciences, Japan) were packed into 7 cm of 150-μm id 360-μm od capillary, resulting in about 2 mg particles packing. Capillary columns were washed with 50% ACN for 10 min at 1.5 μL/min flow rate on a pressure-loader, with 2% ammonia, 20% ACN (elution buffer) and then with a phosphopeptide enrichment (PE) buffer (50% ACN, 14% glycolic acid, and 2% TFA). Peptide samples were reconstituted by adding ACN, glycolic acid, and TFA to make the same composition with PE buffer and loaded onto the column at a 1 μL/min flow rate. The column was then washed with PE buffer for 10 min and with 50% ACN 2% TFA at 1.5 μL/min. The bound peptides were eluted with elution buffer for 5 min at 1.5 μL/min, immediately acidified with 5 μL of 5% formic acid, and then stored at −80°C until used.
2.4. Mass spectrometry
LC-MS/MS analyses of trypsin-digested peptides or enriched phosphopeptides were carried out using a Thermo Fisher Scientific linear ion trap (LTQ/Orbitrap XL) in replicates. Briefly, peptides (1–2 μg) were first loaded onto a trap cartridge (Zorbax 300SB-C18, 5 μm, 0.3x5mm, Agilent) at a flow rate of 2 μL/min. Trapped peptides were then eluted onto a reversed-phase PicoFrit column (Betabasic 5 μm C18, 150 Å, 10 cm bed length, 360 od/75 id, New Objective) using a linear gradient of ACN (2–35%) containing 0.1% formic acid. The duration of the gradient was 80 min at a flow rate of 0.25 μl/min, followed by 80% ACN washing for 5 min. The eluted peptides were sprayed into the LTQ-Orbitrap via a nanospray ion source. The spray voltage and ion transfer tube temperature were set at 1.8 kV and 180°C, respectively. The data-dependent acquisition mode was enabled, and each survey MS scan was followed by four MS/MS scans with dynamic exclusion option on. Full-scan MS survey spectra (m/z 300–2,000) in profile mode were acquired in the Orbitrap (resolution: 60,000; AGC target: 5E+05; max. fill time: 500 ms). The four most intense peptide ions from the survey scan were fragmented by collision-induced dissociation (normalized collision energy: 35%; activation Q: 0.250; and activation time: 30 ms) in the LTQ (AGC target: 1E+04; max. fill time: 200 ms). For TiO2-enriched samples, multistage activation (neutral losses of 97.98, 48.99, 32.66, and 24.50 m/z) was enabled. Precursor ion charge state screening was used to reject unassigned charge states. The dynamic exclusion list was restricted to a maximum of 500 entries with a maximum exclusion duration of 90 s and a relative mass tolerance window of ±10 ppm.
2.5. MS data analysis
Raw data was analyzed using the MaxQuant software environment (version 1.1.1.6.) [15, 16]. Retention time dependent mass recalibration was applied and peak lists were searched against a database containing all 87,061 entries from the International Protein Index human protein database (http://www.ebi.ac.uk/IPI/IPIhelp.html) version 3.68 and 262 frequently observed contaminants as well as the reversed sequences of all entries. Database searches were performed with the following settings. Precursor and fragment ion peaks were searched with mass tolerance of 7 ppm and 0.5 Da, respectively. Enzyme specificity was set to trypsin/P. Up to two missed cleavages were allowed and only peptides with at least 6 amino acids in length were considered. Carbamidomethylcysteine and oxidation on methionine were set as fixed and variable modifications, respectively. For phosphopeptide samples, phosphorylation of serine, threonine, and tyrosine were set as variable modifications. Using a decoy database strategy, peptide identifications were accepted based on their posterior error probability (PEP), until less than 1% reverse hits were retained in the list. Phosphorylation site assignments were performed by the PTM scoring algorithm implemented in MaxQuant. Phosphorylation site assignments were classified as class I sites in case of a localization probability of at least 0.75 and a score difference of at least 5 to the second most likely assignment [17].
2.6. Dataset analysis
All SILAC ratios from MaxQuant were converted to log2-scale and the dataset’s mean (μ) and standard deviation (σ) were calculated with SigmaPlot v11.0. The correlation curve of measured vs. pre-defined mixed ratios and its coefficients were obtained by linear regression (SigmaPlot) and the dataset’s standard deviations were used for error bar calculations for each data point
3. Results
Quantitative proteomic studies of control (non-treated) human neuroblastoma BE(2)-M17 cells using SILAC in our laboratory demonstrated that a majority of log2-transformed H/L ratios were often below zero (i.e. H/L < 1). This was caused by a high arginine-to-proline conversion and incomplete isotope labeling (Supplemental Fig. S1 and S2). With eight division cycles (7 days) in DMEM SILAC media, these cells reached 92% labeling efficiency along with the presentation of multiple satellite peaks of proline-containing peptides. As demonstrated by others [9], arginine-to-proline conversion with the inclusion of additional proline (200 mg/L) was effectively attenuated. With such a proline inclusion, the difference of labeling efficiency between proline-containing and non-containing peptides in our study was only 0.5% in 7-day culture sample. However, labeling efficiency was not further improved with any subsequent increases in proline concentration or more division cycles.
3.1. A theoretical calculation of SILAC ratio
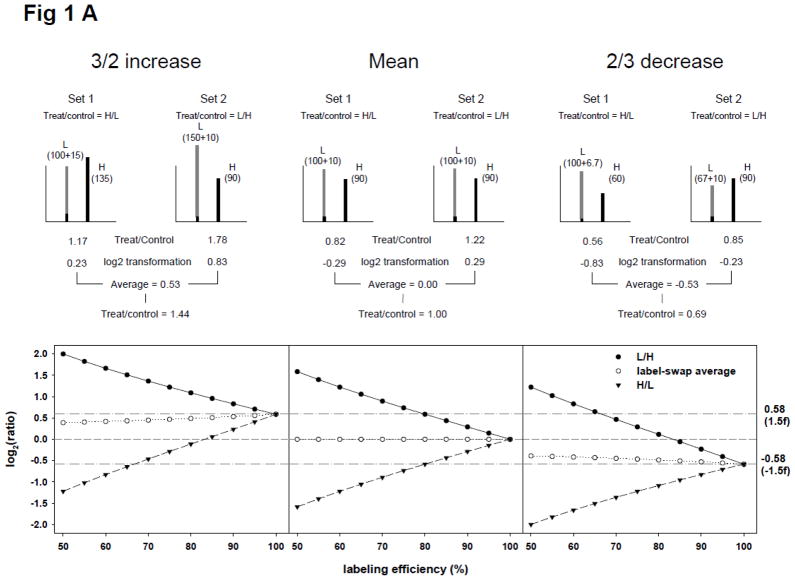
To examine the effect of labeling efficiency and arginine-to-proline conversion on experimental SILAC ratios, we initially calculated theoretical ratios based on a hypothesis that all proteins in H condition were labeled at the same efficiency and without any arginine-to-proline conversion. With a 90% labeling efficiency, peptides in H condition would therefore have a 10% proportion of light peptides, which contribute to the signal of light ions in a SILAC doublet so that an H/L mean ratio would become 0.82 (= 90/(100+10)), and not 1.00 (set 1 of upper middle ‘mean’ panel in Fig. 1A). If a specific protein expression increase of 3/2 by a specific treatment occurred, the 10% proportion of light peptides in the treated H condition would also increase, which would subsequently decrease the H/L ratio to 1.17 (=135/(100+15); set 1 of upper left ‘3/2 increase’ panel in Fig. 1A). In the case of an experimentally-induced 2/3 expression decrease, a resultant H/L ratio of 0.56 (= 60/(100+6.7)) would occur (set 1 of upper right ‘3/2 decrease’ panel in Fig. 1A). Overall, light peptides resulted from incomplete labeling of H condition caused a log2-transformed H/L ratio to shift towards the negative side. This negative shift was significant below conditions with only 80% labeling as the mean log2-transformed ratio would be −0.58 (ratio, 2/3) which is often used as a cut-off for significant protein down-regulation in studies. This negative shift may be completely opposite in a label-swap replicate in which the light condition was treated (set 2 in Fig. 1A). As seen in Fig. 1A, all the calculated ratios of mean, 3/2 increase, and 2/3 decrease, in various labeling efficiencies shift to the negative side for H/L (treated/control: set 1) and to the positive side for L/H (treated/control: set 2). Interestingly however, if the treated/control ratios were averaged using counterparts from label-swap replicates, the resultant values were consistently closer to ideal experimental ones.
Figure 1.
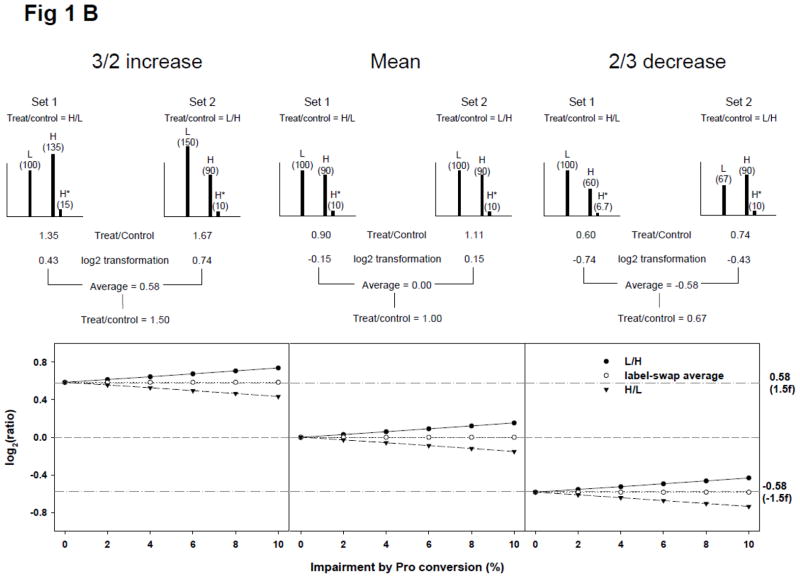
Theoretical calculation of SILAC ratios. (A) Effect of incomplete isotope labeling on SILAC ratios in a practical cell stimulation paradigm. In a theoretical situation in which proteins in the heavy condition (H) in set 1 and the light condition (L) in set 2 (labeled with isotope to 90%) were treated with a stimulus, and the expected ion intensities of SILAC doublet of a peptide are shown in the upper panel. The theoretical data demonstrates that ion intensity of light peptide in SILAC doublet increased by the addition of unlabeled light peptide from H. The ratios of mean, 3/2 increase, and 2/3 decrease in set 1 were calculated to be lower than expected and ratios in set 2 were calculated to be higher than expected. These errors however would be corrected by averaging label-swap replicate SILAC experiments. The lower panel demonstrates the effect of label-swap averaging on SILAC ratios with various labeling efficiencies; (B) Effect of arginine-to-proline conversion on SILAC ratios. The arginine-to-proline conversion process reduces the ion intensity of heavy state entities by generating satellite peaks containing heavy proline. We posited that proteins in H were completely labeled with isotope but ion intensities of proline-containing heavy peptides were reduced by 10% through the arginine-to-proline conversion process. The upper panel demonstrates how label-swap replication could solve the conversion errors. The lower panel demonstrates the effect of arginine-to-proline conversion on SILAC ratios with various conversion percentages. The value of log2(1/1) was shown with dash line and ones of log2(3/2) and log2(2/3) were shown with dashed-dot lines.
Next, we examined the effect of arginine-to-proline conversion on the same theoretical SILAC ratios shown in Fig. 1A, with a hypothesis that proteins were completely labeled and 10% of heavy peptide in H condition was impaired by the conversion, resulting in a 10% decrease of heavy ion intensity. Some satellite peaks of heavy ions would be observed in a SILAC doublet but these peaks would not be considered for ratio calculation in most SILAC applications (upper panel of Fig. 1B). With a 10% reduction of heavy ion intensity, log2-transformed H/L (treated/control) ratios of mean, 3/2 increase, and 2/3 decrease were decreased slightly (negative shift), but the shifting extent was smaller than that of 90% incomplete labeling and the averaged ratios of label-swap replicates were exactly the same as the ‘ideal’ experimental values. Effects of arginine-to-proline conversion on SILAC ratios are shown in the lower panel of Fig. 1B, for various conversion percentages. Taken together, treated/control ratio averaging of label-swap replicates would be effective in compensating for apparent SILAC errors by incomplete labeling and arginine-to-proline conversion.
3.2. Linear relationship of mixed and measured ratios in multiple experimental conditions
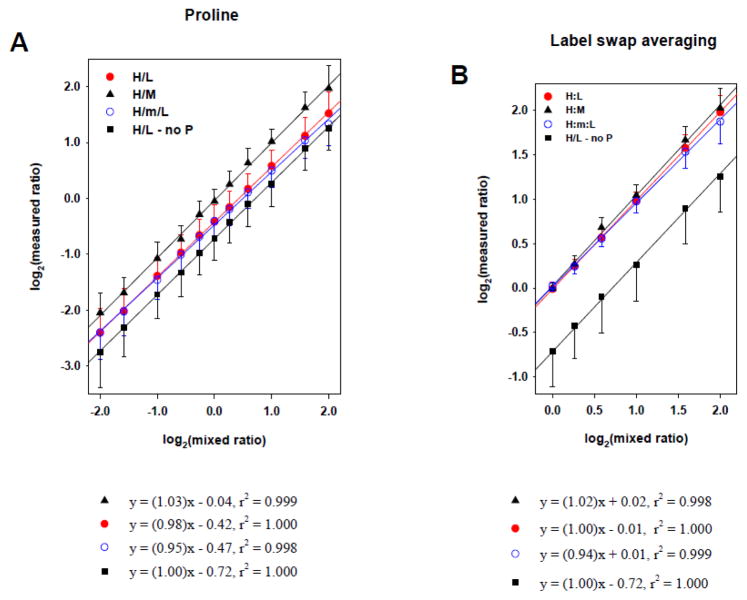
To examine how actual experimental errors are generated by incomplete labeling and/or arginine-to-proline conversion, we investigated the relationship between measured ratios and pre-defined mixed ratios in various experimental conditions. Neuroblastoma BE(2)-M17 cells could be labeled up to 92% and showed strong arginine-to-proline conversion in SILAC DMEM media. Proteins from different isotopic labeling conditions were mixed in 4:1, 3:1, 2:1, 1.5:1, 1.2:1, 1:1, 1:1.2, 1:1.5, 1:2, 1:3, and 1:4 ratios. Dataset means of each mixing ratio scenario (for the multiple labeling conditions) were used for linear regression with its standard deviation as error bar (Fig. 2). In Fig. 2A (“H/L – no P”), protein mixes from L and H conditions in which cells were cultivated with proline-deficient normal DMEM demonstrated an accurate linear relationship between measured and mixed ratios but showed strong experimental error, of negative shift, in which 1/1 ratio was measured as 3/5. Proline addition reduced this negative shift by 40% but still retained significant experimental errors (H/L in Fig. 2A), which were thought to result mainly from incomplete labeling. To avoid the compounding effect of incomplete labeling on ratio correlation, we also examined the H/M ratio in SILAC mixes with only labeled conditions (i.e. H and M) and without L condition. As a peptide in M and H conditions would have the same labeling efficiency and be affected by the same conversion rate, the H/M ratios were expected to be ideally the same with mixed ratios. From the correlation, the negative shift of H/M was indeed significantly reduced (H/M in Fig. 2A), which confirmed that the measurement errors of negative shift of H/L dataset in Fig. 2A were caused by incompletely labeling. We examined H/L ratios in SILAC triplet design (H/M/L in Fig. 2A) because the additional M condition could introduce more complexity. These were measured a little smaller than one in SILAC doublet (without M condition) especially in high ratios, which was likely caused by the additional supply of light ions from M condition by incomplete labeling. With these results, it was concluded that SILAC experiments were very vulnerable to incomplete labeling and arginine-to-proline conversion and the prevention of arginine-to-proline conversion alone was not enough to achieve reliable analysis if incomplete labeling was involved.
Figure 2.
Correlation between measured and pre-defined mixed ratios in various SILAC mixtures. Neuroblastoma BE(2)-M17 cells were cultured in SILAC M (K4R6) or H (K8R10) DMEM media for 7 days, which showed 92% averaged incorporation of isotopic amino acids. Proteins of each condition were mixed in 4:1, 3:1, 2:1, 1.5:1, 1.2:1, 1:1, 1:1.2, 1:1.5, 1:2, 1:3, and 1:4, and their measured ratios were compared with mixed ones. (A) Effect of additional proline on ratio correlation. Proteins from the H condition (H DMEM media without proline) were mixed with ones from the L condition (■). With additional proline in media, proteins from L and H conditions (●) or M and H (▲) were mixed. And also proteins from L and H conditions were mixed in the presence of M labeled proteins which were half of total proteins of L and H mix (○). Their measured ratios correlated well with mixed ones but resulted in negative shift which was the strongest in L and H mix without proline (■) and the smallest in M and H mix with proline (▲); (B) Effect of label-swap averaging on ratio correlation. Measured ratios from L and H mix (●), L and H mix with additional M (○), M and H mix (▲), and L and H mix without proline (■) were averaged in quantified peptides common in both label-swap replicates. All measured ratios correlated well with pre-defined mixed ones without any significant shift. The results of linear regression were shown in the below of each graph and error bars of data point represent standard deviation of dataset of each mix.
Next, we investigated how the ratio averaging of label-swap replicates affected the relationship between measured and pre-defined mixed ratios. Each peptide quantitatively identified at both label-swap replicates (as example, 4/1 = H/L in one set and 1/4 = L/H in the other set) was considered to calculate averaged ratio. Mean and standard deviation of averaged dataset were used for linear regression as mentioned above, in which the proline was added to prevent the arginine-to-proline conversion so as to clarify the effect of label-swap averaging on the negative shift (Fig. 2B). As shown in H:L correlation in Fig. 2B, the label-swap averaging resulted in good correlation with similar linearity and negligible negative shift. Furthermore, the label-swap averaging demonstrated its effectiveness on reducing the negative shift in all the label conditions tested for SILAC mixing (Fig. 2B). Interestingly, as shown in Table 1, this simple process proved to be effective with experiments without additional proline (so in the presence of high arginine-to-proline conversion). Taken together, all the correlations between measured and pre-defined mixed ratios from different experimental conditions retained a good linear relationship, but incomplete labeling and/or arginine-to-proline conversion affected ratio accuracy. The ratio averaging of label-swap replicates was able to solve these experimental errors, allowing for generation of more reliable SILAC ratios. However, it was observed that the measured ratios from label-swap replicates were a little smaller than the real mixed ones, especially for the higher ratios. In case of the SILAC triplet, the greater inherent complexity of incomplete labeling and conversion was induced by the additional M condition and thus the calculated ratios became smaller.
Table 1.
Correlation between measured and pre-defined mixed ratios in various SILAC mixtures.
| SILAC | proline | label-swap | linear regression *
|
st.dv. of dataset | |
|---|---|---|---|---|---|
| slope | shift | ||||
| H/L (with M) | − | − | 1.00 | −0.72 | 0.38 – 0.63 |
| + | − | 0.98 | −0.42 | 0.27 – 0.42 | |
| − | + | 0.99 | 0.02 | 0.07 – 0.22 | |
| + | + | 1.00 | −0.01 | 0.07 – 0.19 | |
| + | − | 0.95 | −0.47 | 0.28 – 0.49 | |
| + | + | 0.94 | 0.01 | 0.06 – 0.25 | |
|
| |||||
| H/M | − | − | 1.00 | −0.09 | 0.22 – 0.42 |
| + | − | 1.03 | −0.04 | 0.22 – 0.41 | |
| − | + | 0.98 | 0.04 | 0.06 – 0.24 | |
| + | + | 1.02 | 0.02 | 0.06 – 0.22 | |
log2(mean of measured dataset) = (slope) × log2(mean of mixed ratio) + (shift)
3.3. Analysis of a ligand-regulated proteome and phosphoproteome using label-swap replication
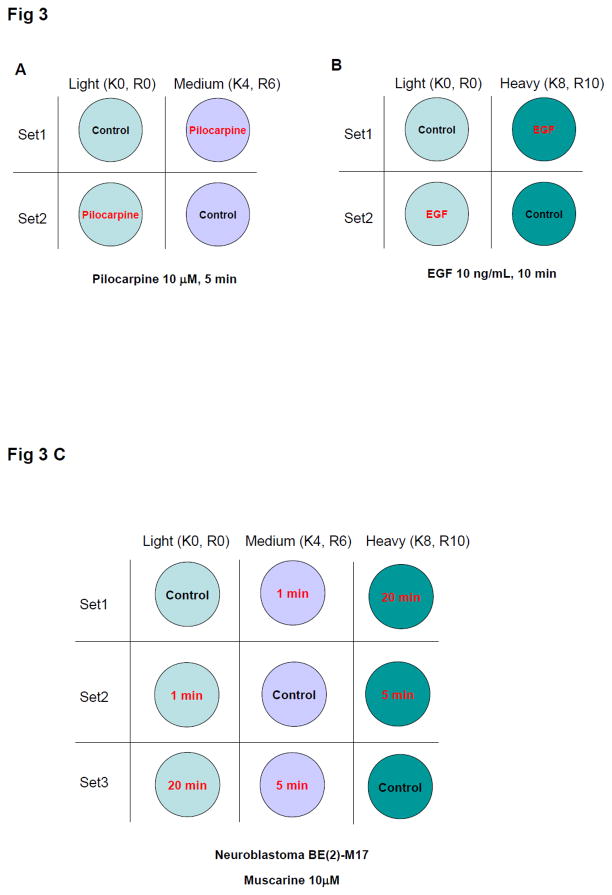
To examine how the ratio averaging of label-swap replicates affected SILAC experimentation, ligand-receptor signaling was investigated. The ligand-receptor binding event often triggers a rapid cellular series of multiple protein phosphorylation events. Receptor-mediated signaling events can be readily detected within minutes after ligand binding; therefore protein phosphorylation is likely to occur without substantial changes in gross protein expression [17–20]. Human neural BE(2)-M17 cells were stimulated with the growth factor receptor ligand EGF (epidermal growth factor: 10 ng/mL, 10 min) or the muscarinic acetylcholine receptor agonist pilocarpine (10 μM, 5 min). The H condition was used for the label-swap replication for EGF treatments and the M condition was applied to the pilocarpine protocol (Fig. 3A and B). Gross expression levels of most proteins were expected not to be altered in the short experimental time periods (5–10 min) by the ligand treatments; but cellular signaling, represented by protein phosphorylation, would alter rapidly and differentially. EGF is known to rapidly activate multiple cellular signaling pathways, and in doing so, induce phosphorylation of many intracellular proteins [17, 18]. The responses of muscarinic receptors to stimulatory ligands are controlled by the nature of the agonist molecule as well as their in vivo environment [21, 22]. In M17 cells, pilocarpine, known as a partial muscarinic receptor agonist, caused only a minimal extracellular signal-regulated kinase (ERK1/2) phosphorylation, while a full muscarinic receptor agonist, e.g. muscarine, triggered a more robust ERK1/2 phosphorylation response (Supplementary Fig. S3). We employed both strong (EGF) and weak (pilocarpine) signaling stimuli to demonstrate that our methodology veracity was independent of the stimulatory modulus.
Figure 3.
Experimental design of ligand treatment using label-swap. Neuroblastoma BE(2)-M17 cells were treated with 10 μM pilocarpine for 5 min (A), with 10 ng/mL EGF for 10 min (B), or with 10 μM muscarine for 1, 5, and 20 min (C). In replicated sets, labeled conditions of treatment and control were swapped in both SILAC doublet and triplet.
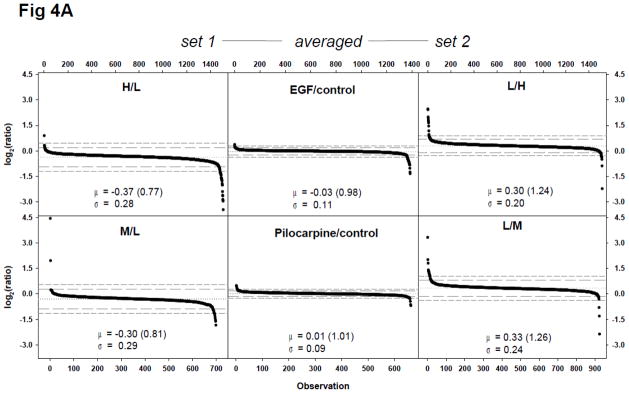
Equal amounts of proteins were mixed for the SILAC doublet experiments (EGF, pilocarpine: Fig. 3) using label-swap design; with trypsin-digested peptides and phosphopeptides analyzed without further fractionation. Based on theoretical calculations, the mean of log2-transformed ratios of a proteome dataset with 92% labeling efficiency was expected to be −0.23 (0.85 in ratio) for set 1 (H/L = treated/control), 0.23 (1.17 in ratio) for set 2 (L/H = treated/control), and zero for averaged ones (Fig. 1A). Proteomic analyses demonstrated similar or marginally more shifting of the dataset mean; −0.37 (0.77 in ratio) for set 1 and 0.30 (1.24 in ratio) for set 2 of EGF experiment and −0.30 (0.81 in ratio) for set 1 (M/L = Pilocarpine(Pilo)/control) and 0.33 (1.26 in ratio) for set 2 (L/M = Pilo/control) of the pilocarpine experiment (Fig. 4A). In addition, the majority of measured ratios in the set 1 experiment were below zero and the ratios in the set 2 experiment were above zero (Fig. 4A). However, the ratio averaging of label-swap replicates resulted in −0.03 (ratio of 0.98) for the EGF experiment and 0.01 (ratio of 1.01) for the pilocarpine experiment. The proteome datasets’ standard deviation was reduced more than 50% with the ratio averaging process in both treatments.
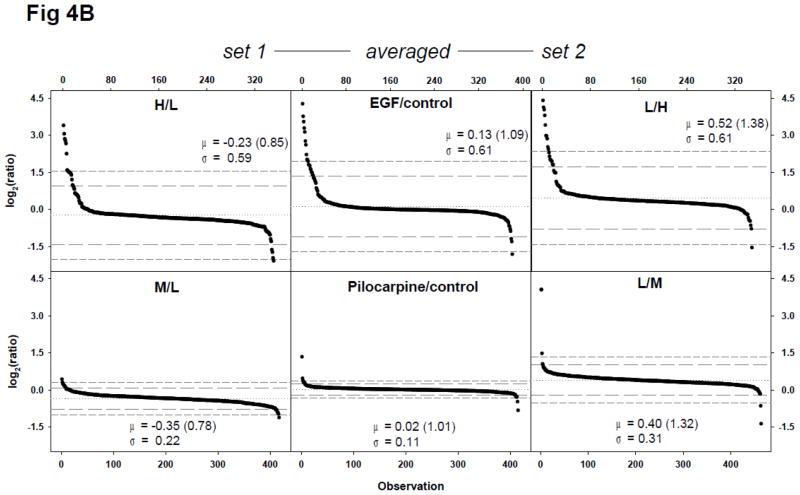
Figure 4.
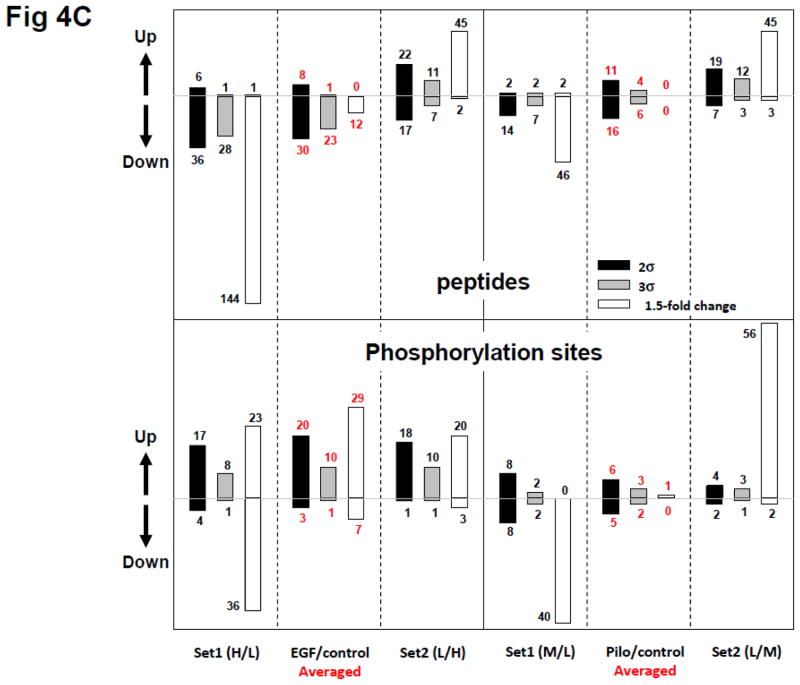
Quantitative proteomic and phosphoproteomic analyses of EGF or pilocarpine treatment of neural cells using a label-swap SILAC doublet. Neuroblastoma BE(2)-M17 cells were treated with 10 ng/mL EGF for 10 min, or with 10 μM pilocarpine for 5 min, and trypsin-digested peptides and enriched phosphopeptides were analyzed via LC-MS/MS without fractionation. (A) Scatter plot of log2-transformed ratio of individual quantified peptides in set 1 and 2 and label-swap averaged dataset. From EGF-treated samples, 1508 peptides were quantified in set 1 and 1514 ones in set 2. Of these peptides, 1386 ones were commonly quantified. From pilocarpine-treated samples, 698 peptides were quantified in set 1 and 924 ones in set 2, with 662 common in both sets; (B) Scatter plot of log2-transformed ratio of individual class I phosphorylation sites in set 1 and 2 and label-swap averaged dataset. From EGF-treated samples, 351 class I phosphorylation sites were quantified in set 1 and 348 sites in set 2. Of these sites, 381 those which were categorized as class I site in at least one set were commonly quantified in both sets. From pilocarpine-treated samples, 417 class I phosphorylation sites were quantified in set 1 and 464 sites in set 2. Of these sites, 416 those which were categorized as class I site in at least one set were commonly quantified in both sets. Values for mean (μ) and standard deviation (σ) are shown in each plot and indicated with a dotted line for mean, a long dashed line for 2σ, and a short dashed line for 3σ; (C) Significant protein and phosphorylation site alterations. Numbers of significantly up- or down-regulated peptides or phosphorylation sites in each dataset with three cut-offs, ±2σ (black bars), ±3σ (grey bars), and ±1.5-fold change (white bars) are depicted.
Phosphoproteome analyses of EGF and pilocarpine stimulation, as expected, demonstrated wider ratio distributions (Fig. 4B). Interestingly, the standard deviations of the pilocarpine phosphoproteome datasets were similar to those of the non-phospho proteome analysis, while the EGF datasets demonstrated considerably larger standard deviations in phosphoproteome analysis and did not exhibit the log2(ratio) shrinkage trend between set 1 and 2 and label-swap averaged datasets (Fig. 4B).
We also examined significant alterations in all the peptide datasets with three statistical or magnitude cut-offs, ±2σ, ±3σ, and ±1.5-fold change (upper panel of Fig. 4C). Using the statistical cut-offs (±2σ, ±3σ), relatively little difference in the representation of ‘changed’ expression was seen between set 1 or set 2 data for both EGF and pilocarpine treatment. There were minimal changes in total protein expression when averaging was used to finalize the data (red numbers – upper panel Fig. 4C). However, if the 1.5 fold cut-off was used (white bars) without averaging across the label swap, large expression change errors were apparent between set 1 and set 2 data for both EGF and pilocarpine treatment. Despite these considerable differences (using a 1.5 fold cut-off) in set 1 and set 2 data, a minimal protein expression change was obtained with averaging, similar to those with the ±2σ or ±3σ cut-offs. If significantly altered peptides (1.5-fold change cut-off) were false positives, 92% (upper left panel of Fig. 4C, from set 1 to averaged, 92% = (145 − 12)/145) and 74% (upper left panel of Fig. 4C, from set 2 to averaged, 74% = (47 − 12)/47) of these in EGF dataset were regarded to be corrected. Interestingly, all significantly altered peptides (potentially false positives) in pilocarpine dataset were eliminated by our method (upper right panel of Fig. 4C). This demonstrates that even with large data disparities, label-swap averaging is able to effectively correct experimental errors.
When we similarly investigated the EGF- or pilocarpine-induced protein phosphorylation data (Fig. 4C, lower panel), we again demonstrated that the larger errors associated with the 1.5 fold cut-off (white bars), as opposed to the ±2σ or ±3σ cut-offs, could be effectively removed by label-swap averaging. Based on these observations and the nature of the EGF and pilocarpine effects on neural M17 cells, we concluded that just one set of SILAC analysis could lead to significant quantification errors and ratio averaging of label-swap replicates could be a simple solution for accurate quantification. It could also be proposed that if SILAC ratios were accurately measured, significant alterations could be selected based on fold-change combined with label-swap averaging.
3.4. SILAC triplet using label-swap replication
Compared to the relatively simple doublet experiments, SILAC triplet can introduce extra levels of measurement errors because light peptides can be supplied from both M and H conditions during incomplete labeling. If arginine-to-proline conversion is also involved, experimental errors are likely to be more complicated by additional satellite peaks from the M condition. We therefore examined how the ratio averaging of label-swap replicates affected SILAC ratios in a triplet experiment. We compared four different experimental conditions including a control set with three SILAC mixtures. We applied the label-swap replication to a SILAC triplet in which the temporal response of M17 cells to a more efficacious muscarinic acetylcholine receptor agonist, muscarine, were analyzed with an experimental design containing 1, 5, and 20 min treatments of 10 μM muscarine (Fig. 3C). Proteomic and phosphoproteomic analyses of each SILAC mix resulted in about 830 peptides and 450 class I phosphorylation sites, respectively. Of these, 78% peptides and 75% phosphorylation sites were repetitively detected in all three experimental mixtures (Fig. S4A).
The datasets’ mean and standard deviation are shown in Table 2. Diagrammatical plots of their ratio distributions are represented in Supplementary Fig. S4B and S4C. Data set 1 or set 2 of the 1 min and 20 min time points exhibited significant negative or positive shift, respectively, which resulted in a high level of significant alterations using the 1.5-fold change cut-off limit in both proteome and phosphoproteome datasets. The ratio shift of the 5 min time point dataset was the smallest of the three time points studied, as unlabeled light peptides from M and H conditions were excluded for the resultant ratio measurement. However, as seen in Fig. S4B/C, ratio averaging rendered almost all ratios to be between 0.67 and 1.50, demonstrating no significant alterations through all the time points, while several significant alterations were selected by statistical cut-offs (±2σ, ±3σ) because of very narrow distribution.
Table 2.
Statistical analysis of proteome and phosphoproteome dataset of temporal response of neural BE(2)-M17 cells to the muscarinic receptor ligand, muscarine.
| dataset | labeling | mean
|
st.dv. | |
|---|---|---|---|---|
| log2(ratio) | ratio | |||
| Peptides | ||||
| 1 min/control |
label-swap average | −0.10 | 0.94 | 0.10 |
| M/L | −0.51 | 0.70 | 0.33 | |
| L/M | 0.32 | 1.25 | 0.35 | |
| 5 min/control |
label-swap average | 0.00 | 1.00 | 0.09 |
| H/M | −0.10 | 0.93 | 0.20 | |
| M/H | 0.10 | 1.07 | 0.25 | |
| 20 min/control |
label-swap average | 0.02 | 1.01 | 0.09 |
| H/L | −0.38 | 0.77 | 0.30 | |
| L/H | 0.43 | 1.35 | 0.32 | |
|
| ||||
| phosphopeptides | ||||
| 1 min/control |
label-swap average | −0.05 | 0.97 | 0.39 |
| M/L | −0.49 | 0.71 | 0.39 | |
| L/M | 0.38 | 1.30 | 0.48 | |
| 5 min/control |
label-swap average | 0.08 | 1.06 | 0.54 |
| H/M | −0.05 | 0.97 | 0.60 | |
| M/H | 0.20 | 1.15 | 0.56 | |
| 20 min/control |
label-swap average | 0.08 | 1.05 | 0.42 |
| H/L | −0.37 | 0.77 | 0.44 | |
| L/H | 0.52 | 1.43 | 0.49 | |
Phosphoproteome datasets in response to muscarine stimulation demonstrated similar trends but much larger standard deviations indicated significant alterations (mostly up-regulated). Among these, the Tyr187 site of ERK 1 was well known to be actively regulated by muscarinic receptor activation by its cognate ligand. Interestingly, the proteome dataset’s standard deviations were reduced by ratio averaging but it did not occur with the phosphoproteome dataset (Fig. S4), which was reminiscent of the EGF phosphoproteome data (Fig. 4). Therefore the ratio averaging of label-swap replicates was not only able to correct experimental errors of SILAC but also retained the specific functional characteristics intrinsic to selective biological response datasets.
The ratio averaging of label-swap replications also affected the proper choice of required cut-offs to indicate significant alteration events. The statistical cut-offs showed relatively the same numbers of alterations within a treatment, regardless of set coordinations and their averaged ratios, but the fold change cut-off demonstrated dramatic difference between both set coordinations and their averaged ratios, especially in the respective phosphoproteome (EGF, pilocarpine) datasets. In EGF treatment, 59 phosphorylation sites out of 351 ones in set 1 and 23 sites out of 348 ones in set 2, were regarded as significant alterations with a 1.5-fold change cut-off and 36 phosphorylation sites out of 381 ones in the ratio-averaged dataset (Fig. 4C). As shown in Fig. 4C, with pilocarpine treatment, just two phosphorylation sites out of 415 ones were regarded as significant alterations with the 1.5-fold change cut-off in the ratio-averaged dataset. This was observed again in SILAC triplet experiments of the temporal phosphoproteomic response to muscarine treatment. In the proteome dataset, all averaged ratios of three time points were within 1.5-fold change, however using the ±3σ cut-off, 2-4 protein groups came out of about 130 ones as significant alterations. In the phosphoproteome dataset, about 20 phosphorylation sites were outside of 1.5-fold change and these increased from 15 to 28 ones across the time course. When applying a ±3σ cut-off, this resulted in the selection of only slightly fewer phosphorylation sites, of which the number was relatively constant during the total 20 minute time course. Taken all together, the ratio averaging of label-swap replicates demonstrated the effective correction of SILAC experimental errors.
4. Discussion
The incidence of experimental errors can often be problematical for the interpretation of SILAC data in the context of cellular signaling functions. Our work has attempted to attenuate one of the major sources of experimental errors in SILAC applications to facilitate more accurate quantification. Our theoretical calculations of SILAC ratios in several circumstances revealed the quantification errors by the addition of light peptides from the heavy condition and reduction of heavy peptide ions via arginine-to-proline conversion. Interestingly, the errors incurred by incomplete labeling were calculated to be bigger than those introduced by the amino acid conversion process, as unlabeled light peptides from the heavy condition were subjected to quantification (thus causing errors), but not the heavy proline satellite peaks. In performing an archetypical SILAC doublet experiment, such as H/L (treated/control), all measured ratios would include some significant errors related to both the extent of labeling efficiency and amino acid conversion rates. Application of suitable statistical cut-offs may be able to select some true-positive alterations but the reliability of measured ratios would still be highly dependent on the vagaries of labeling efficiency. Through a facile process, i.e. ratio averaging of label-swap replicates, attenuation of these SILAC errors, by compensation of each of the aforementioned sources of experimental inaccuracy, can be achieved. Despite considerable efforts being made to solve issues of arginine-to-proline conversion in SILAC applications, it still is a problematic aspect for most organisms or cell lines. Surprisingly, errors induced by the amino acid conversion process were shown to be completely compensated for through the ratio averaging using our theoretical calculations. However, high labeling efficiency was important as incomplete labeling could cause false-negatives as the averaged ratios of 3/2 increase and 2/3 decrease were brought closer to zero with low labeling efficiency, as demonstrated in the lower panel in Fig. 1A.
In our experiments it was not feasible to achieve above ≥93% labeling with the neural BE(2)-M17 cells, even with proline addition of the growth media or longer cultivation time. As the difference of labeling efficiency between proline-containing peptides and non-proline-containing peptides was just 0.5% from the labeling test analyses, it was concluded that the arginine-to-proline conversion occurred at a very low rate in the presence of additional proline. As extended cultivation time was not able to increase labeling %, this suggested that there were other, unknown and complicating, factors responsible for incomplete protein labeling. It has been postulated that one of the common complicating factors is the presence of serum proteins in the cell growth media. Serum proteins can be digested by cellular proteases with the resultant amino-acids then available for cellular protein synthesis. For example, if the percentage of amino-acid supply from serum-protein digestion would be 8%, it could easily account for the 92% labeling efficiency we observed in M17 cells. Testing this hypothesis however, with low-serum culture conditions, is fraught with the introduction of additional biological variation, e.g. most cells demonstrate different growth rates and significantly altered cellular properties in different serum environments. In normal cell culture conditions, certain cells like BE(2)-M17 are expected to show both incomplete labeling and amino acid conversion, resulting in significant quantification errors. As summarized in Table 1, our method corrected quantification errors by almost 97% in SILAC H/L doublet (based on the shift of linear regression) in this situation. Therefore a simple experimental strategy, such as our ratio averaging of label-swap replication, could be applied to facilitate the broad acceptability of SILAC to multiple cell lines or organisms by reducing commonly occurring experimental errors. The label swap replication was designed to correct quantification errors in SILAC experiments. Our process necessitates the introduction of a biological replicate rather than just a simple technical replicate. Biological replicates are more likely to introduce identity variability, unlike a technical MS run replicate, but will likely increase the comparability of data between experimental researchers, as only the most consistently-phosphorylated proteins will be accurately quantified and reported. Therefore despite introducing potential variability in identity generation, a more physiologically-relevant datastream may be created from the more reliably identified phosphorylated proteins. Experimenters however should perform careful sample handling to minimize biological variation between label-swap replicates.
The label-swap replication strategy was tested in SILAC doublet and triplet experimental formats in which the ligand-mediated receptor signaling was a good candidate to study. It is known that EGF is able to regulate multiple facets of intermediary cell metabolism through cellular responses involving protein phosphorylation. Due to the rapid nature of the post-translational modifications, a significant change in expression levels is highly unlikely to occur during the same period. It has been demonstrated in recent years that the ligand-mediated activation of G protein-coupled receptors (GPCRs) can also stimulate signaling cascades highly reminiscent of those induced by growth factors such as EGF [23, 24]. However, the interaction of ligands with GPCRs is relatively nuanced, compared to growth factor receptors in that a broad spectrum of ligand signaling efficacy is common. We have demonstrated that the muscarinic receptor agonist with greater intrinsic efficacy, muscarine, was able to trigger a more significant phosphorylation response compared to pilocarpine in the M17 cells. Therefore, it was expected that EGF and muscarine would activate various cellular signaling in a more robust manner than pilocarpine and that these three ligands would not affect total protein expression level in a short time period. Reinforcing the good physiological translational status of our experiments and subsequent data analysis we indeed observed all of these expected subtle pharmacological phenomena, e.g. variable GPCR ligand efficacy. This is important as phosphoproteomic profiling of receptor response patterns is likely to represent an important new field of investigation for rational drug design [25, 26]. With regards to the specific physiological nature of protein phosphorylation in response to EGF, we found increases in phosphorylation of multiple proteins previously detected to be sensitive to EGF stimulation, e.g. microtubule-associated protein 1B [27], stathmin [28] and serine/arginine repetitive matrix protein 2 [29]. In a similar manner we were also able to corroborate, with our datasets, the phosphorylation of proteins also previously associated with muscarinic receptor signaling cascades, e.g. mitogen-activated protein kinase 1 [30], myristoylated alanine-rich C-kinase substrate [31] and heat-shock protein 27kDa [32]. Therefore our identified phosphorylation events appear to be consistent with expected results of experimental receptor stimulation.
From our proteomic and phosphoproteomic analyses, all the experiments of ‘treated/control’ set 1 (H/L or M/L) exhibited ratio distributions represented by a low dataset mean; while the converse in set 2 (L/H or L/M) was typified by high means, as expected by our theoretical calculation. The ratio averaging of all the treatments was therefore likely to compensate for procedural errors as the dataset means became close to 1/1, an ideal value representing no expression change, which is expected to reduce the false positives. As expected from our experimental design, proteome and phosphoproteome datasets showed considerable differences. The standard deviation of phosphoproteome datasets were greater than those of proteome datasets except for the pilocarpine treatment, likely representing the subtle differences in the efficacy of ligand-receptor-mediated control over active cellular phosphorylation events, i.e. pilocarpine is only a partial GPCR agonist. There was a possibility that standard deviation was reduced because ratios from two replicates were averaged, but our phosphoproteome datasets from EGF and muscarine stimulations showed that when significant alterations were induced, the standard deviation of dataset was not much affected by ratio averaging, compared with the experimental conditions supposed to be no alternation (pilocarpine phosphoproteome dataset).
Statistical analyses of large datasets, such as normalization have been successful with respect to overcoming some experimental errors [33], however, simplistic SILAC proteomic analysis with one kind of experimental design (such as heavy over light) could lead to incorrect measured ratios if incomplete labeling or amino acid conversion was actively involved. Our easy to use ratio averaging process of label-swap replicates may represent a readily applicable solution to the experimental errors. We suggest that quantitative proteomics using SILAC should be designed in label-swap replication to obtain ratio data by averaging commonly quantified peptides in both replicates. While our simple process can effect accurate error compensation, alternative solutions for different laboratories still present themselves, e.g. labeling efficiency measurement prior to sample mixing or computational post-data acquisition correction. However, there are still many issues with these alternative approaches. For example, with respect to labeling efficiency measurement, the specific labeling extent and amino-acid conversion profile of individual proteins can be highly variable (especially certain primary cells do not proliferate well to achieve consistent label incorporation for individual proteins) and thus applying an average calculation across all the data may further induce additional errors. While not being the perfect solution, our ratio averaging strategy can provide more accurate dataset generation, with the additional flexibility that it can also be used synergistically with statistical normalization, rather than being a lone alternative to it.
Supplementary Material
Highlight.
Incomplete isotope labeling and amino acid conversion result in significant quantification errors.
Ratio average of label-swap replicates corrected quantification errors.
Proteome and phosphoproteome studies of receptor signaling demonstrated quantification error correction by label-swap.
Acknowledgments
This work was carried out entirely with the support of the Intramural Research Program of the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gygi SP, Rist B, Gerber SA, Turecek F, Gelb MH, Aebersold R. Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat Biotechnol. 1999;17:994–9. doi: 10.1038/13690. [DOI] [PubMed] [Google Scholar]
- 2.Ross PL, Huang YLN, Marchese JN, Williamson B, Parker K, Hattan S, et al. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol Cell Proteomics. 2004;3:1154–69. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- 3.Thompson A, Schäfer J, Kuhn K, Kienle S, Schwarz J, Schmidt G, et al. Tandem mass tags: a novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Anal Chem. 2003;75:1895–904. doi: 10.1021/ac0262560. [DOI] [PubMed] [Google Scholar]
- 4.Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, et al. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1:376–86. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 5.Ibarrola N, Kalume DE, Gronborg M, Iwahori A, Pandey A. A proteomic approach for quantitation of phosphorylation using stable isotope labeling in cell culture. Anal Chem. 2003;75:6043–9. doi: 10.1021/ac034931f. [DOI] [PubMed] [Google Scholar]
- 6.Ong SE, Kratchmarova I, Mann M. Properties of 13C-substituted arginine in stable isotope labeling by amino acids in cell culture (SILAC) J Proteome Res. 2003;2:173–81. doi: 10.1021/pr0255708. [DOI] [PubMed] [Google Scholar]
- 7.O’Quinn PR, Knabe DA, Wu G. Arginine catabolism in lactating porcine mammary tissue. J Anim Sci. 2002;80:467–74. doi: 10.2527/2002.802467x. [DOI] [PubMed] [Google Scholar]
- 8.Bicho CC, de Lima Alves F, Chen ZA, Rappsilber J, Sawin KE. A genetic engineering solution to the “arginine conversion problem” in stable isotope labeling by amino acids in cell culture (SILAC) Mol Cell Proteomics. 2010;9:1567–77. doi: 10.1074/mcp.M110.000208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bendall SC, Hughes C, Stewart MH, Doble B, Bhatia M, Lajoie GA. Prevention of amino acid conversion in SILAC experiments with embryonic stem cells. Mol Cell Proteomics. 2008;7:1587–97. doi: 10.1074/mcp.M800113-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Van Hoof D, Pinkse MW, Oostwaard DW, Mummery CL, Heck AJ, Krijgsveld J. An experimental correction for arginine-to-proline conversion artifacts in SILAC-based quantitative proteomics. Nat Methods. 2007;4:677–8. doi: 10.1038/nmeth0907-677. [DOI] [PubMed] [Google Scholar]
- 11.Ong SE. Unbiased identification of Protein-Bait Interactions Using Biochemical Enrichment and Quantitative Proteomics. Cold Spring Harb Protoc. 2010 doi: 10.1101/pdb.prot5400. [DOI] [PubMed] [Google Scholar]
- 12.Xia Q, Hendrickson EL, Wang T, Lamont RJ, Leigh JA, Hackett M. Protein abundance ratios for global studies of prokaryotes. Proteomics. 2007;7:2904–19. doi: 10.1002/pmic.200700267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schulze WX, Mann M. A novel proteomic screen for peptide–protein interactions. J Biol Chem. 2004;279:10756–64. doi: 10.1074/jbc.M309909200. [DOI] [PubMed] [Google Scholar]
- 14.Park SS, Maudsley S. Discontinuous pH gradient-mediated separation of TiO2-enriched phosphopeptides. Anal Biochem. 2011;409:81–8. doi: 10.1016/j.ab.2010.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–72. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 16.Cox J, Neuhauser N, Michalski A, Scheltema RA, Olsen JV, Mann M. Andromeda – a peptide search engine integrated into the MaxQuant environment. J Proteome Res. 2011;10:1794–805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- 17.Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, et al. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127:635–48. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 18.Maudsley S, Pierce KL, Zamah AM, Miller WE, Ahn S, Daaka Y, et al. The beta(2)-adrenergic receptor mediates extracellular signal-regulated kinase activation via assembly of a multi-receptor complex with the epidermal growth factor receptor. J Biol Chem. 2000;275:9572–80. doi: 10.1074/jbc.275.13.9572. [DOI] [PubMed] [Google Scholar]
- 19.Christensen GL, Kelstrup CD, Lyngsø C, Sarwar U, Bøgebo R, Sheikh SP, et al. Quantitative phosphoproteomics dissection of seven-transmembrane receptor signaling using full and biased agonists. Mol Cell Proteomics. 2010;9:1540–53. doi: 10.1074/mcp.M900550-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schreiber TB, Mäusbacher N, Kéri G, Cox J, Daub H. An integrated phosphoproteomics work flow reveals extensive network regulation in early lysophosphatidic acid signaling. Mol Cell Proteomics. 2010;9:1047–62. doi: 10.1074/mcp.M900486-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Griffin MT, Figueroa KW, Liller S, Ehlert FJ. Estimation of agonist activity at G protein-coupled receptors: analysis of M2 muscarinic receptor signaling through Gi/o, Gs, and G15. J Pharmacol Exp Ther. 2007;321:1193–207. doi: 10.1124/jpet.107.120857. [DOI] [PubMed] [Google Scholar]
- 22.Maudsley S, Martin B, Luttrell LM. G protein-coupled receptor signaling complexity in neuronal tissue: implications for novel therapeutics. Curr Alzheimer Res. 2007;4:3–19. doi: 10.2174/156720507779939850. [DOI] [PubMed] [Google Scholar]
- 23.Maudsley S, Zamah AM, Rahman N, Blitzer JT, Luttrell LM, Lefkowitz RJ, et al. Platelet-derived growth factor receptor association with Na(+)/H(+) exchanger regulatory factor potentiates receptor activity. Mol Cell Biol. 2000;20:8352–63. doi: 10.1128/mcb.20.22.8352-8363.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Davidson L, Pawson AJ, López de Maturana R, Freestone SH, Barran P, Millar RP, et al. Gonadotropin-releasing hormone-induced activation of diacylglycerol kinase-zeta and its association with active c-src. J Biol Chem. 2004;279:11906–16. doi: 10.1074/jbc.M310784200. [DOI] [PubMed] [Google Scholar]
- 25.Maudsley S, Martin B, Luttrell LM. G protein-coupled receptor signaling complexity in neuronal tissue: implications for novel therapeutics. Curr Alzheimer’s Res. 2007;4:3–19. doi: 10.2174/156720507779939850. [DOI] [PubMed] [Google Scholar]
- 26.Martin B, Brenneman R, Golden E, Walent T, Becker KG, Prabhu VV, et al. Growth factor signals in neural cells: coherent patterns of interaction control multiple levels of molecular and phenotypic responses. J Biol Chem. 2009;284:2493–511. doi: 10.1074/jbc.M804545200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoshi M, Nishida E, Inagaki M, Gotoh Y, Sakai H. Activation of a serine/threonine kinase that phosphorylates microtubule-associated protein 1B in vitro by growth factors and phorbol esters in quiescent rat fibroblastic cells. Eur J Biochem. 1990;193:513–19. doi: 10.1111/j.1432-1033.1990.tb19366.x. [DOI] [PubMed] [Google Scholar]
- 28.Ji H, Baldwin GS, Burgess AW, Moritz RL, Ward LD, Simpson RJ. Epidermal growth factor induces serine phosphorylation of stathmin in a human colon carcinoma cell line (LIM 1215) J Biol Chem. 1993;268:13396–405. [PubMed] [Google Scholar]
- 29.Mukherji M, Brill LM, Ficarro SB, Hampton GM, Schultz PG. A phosphoproteomic analysis of the ErbB2 receptor tyrosine kinase signaling pathways. Biochemistry. 2006;45:15529–40. doi: 10.1021/bi060971c. [DOI] [PubMed] [Google Scholar]
- 30.Chadwick W, Zhou Y, Park SS, Wang L, Mitchell N, Stone MD, et al. Minimal peroxide exposure of neuronal cells induces multifaceted adaptive responses. PLoS One. 2010;5:e14352. doi: 10.1371/journal.pone.0014352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haas MS, Dokas LA. Muscarinic receptor- and phorbol ester-stimulated phosphorylation of protein kinase C substrates in adult and neonatal cortical slices. Brain Res Dev Brain Res. 1999;114:89–98. doi: 10.1016/s0165-3806(99)00023-1. [DOI] [PubMed] [Google Scholar]
- 32.Somara S, Gilmont RR, Varadarajan S, Bitar KN. Phosphorylated HSP20 modulates the association of thin-filament binding proteins: caldesmon with tropomyosin in colonic smooth muscle. Am J Physiol Gastrointest Liver Physiol. 2010;299:1164–76. doi: 10.1152/ajpgi.00479.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maudsley S, Chadwick W, Wang L, Zhou Y, Martin B, Park SS. Bioinformatic approaches to metabolic pathways analysis. Methods Mol Biol. 2011;756:99–130. doi: 10.1007/978-1-61779-160-4_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.