Abstract
VarioWatch (http://genepipe.ncgm.sinica.edu.tw/variowatch/) has been vastly improved since its former publication GenoWatch in the 2008 Web Server Issue. It is now at least 10 000-times faster in annotating a variant. Drastic speed increase, through complete re-design of its working mechanism, makes VarioWatch capable of annotating millions of human genomic variants generated from next generation sequencing in minutes, if not seconds. While using MegaQuery of VarioWatch to quickly annotate variants, users can apply various filters to retrieve a subgroup of variants according to the risk levels, interested regions, etc. that satisfy users’ requirements. In addition to performance leap, many new features have also been added, such as annotation on novel variants, functional analyses on splice sites and in/dels, detailed variant information in tabulated form, plus a risk level decision tree regarding the analyzed variant. Up to 1000 target variants can be visualized with our carefully designed Genome View, Gene View, Transcript View and Variation View. Two commonly used reference versions, NCBI build 36.3 and NCBI build 37.2, are supported. VarioWatch is unique in its ability to annotate comprehensively and efficiently millions of variants online, immediately delivering the results in real time, plus visualizes up to 1000 annotated variants.
INTRODUCTION
Over the past few years, the throughput of the next generation sequencing (NGS) technologies have been exponentially increased to a massive scale, greatly changing the face of genomic research and making post-sequencing data analysis tremendously difficult. This technology improvement calls for powerful and handy bioinformatics tools that can process with high performance the NGS data, such as genomic variants, as well as satisfy analysis features to facilitate research. Many genomic variants annotation online tools published (1–4) or not published like SeattleSeq Annotation (http://snp.gs.washington.edu/SeattleSeqAnnotation134/) and offline tools (5–8) are available, but VarioWatch is unique in its ability to annotate comprehensively and efficiently millions of variants online, immediately delivering the results in real time, plus visualizes up to 1000 annotated variants. Based on GenoWatch (9), serving since 2006 and published in the 2008 Web Server issue, VarioWatch was developed with the aim to offer the research community extremely efficient online annotation service of human genomic variants in the NGS era.
VarioWatch has two major improvements. One is speed and the other is comprehensiveness. Regarding speed, the superseded GenoWatch relied on web robots to retrieve data from many public domain websites, such as NCBI (10–12), UniProt (13), KEGG (14) and GO (15), to annotate bulks of variants. It always provided the up-to-date annotations, and this strategy was sufficient before NGS prevailed. Due to slow responses from the source websites, GenoWatch failed to cope with massive online annotation. To solve the problem, we changed our approach by replacing the idea of always providing the most up-to-date information from the Internet with the idea of providing information from frequently updated local databases. By constructing local databases, we increased the annotating speed to at least 10 000-times faster and kept data integrity better by completely avoiding source information retrieval through internet connection and the instability of external web sites. Now that the system is re-structured, re-programmed and fine-tuned, millions of variants can be analyzed and downloaded in minutes, if not seconds, in CSV format with MegaQuery, and up to 1000 variants can be easily visualized and browsed. In addition, we provided filters in MegaQuery to help users narrow down candidate variants and expedite their research.
On top of speed increase, VarioWatch also offers more comprehensive analysis. In contrast to GenoWatch annotating only known SNPs, VarioWatch analyzes both known SNPs and novel variants. By incorporating features similar to FANS (16), VarioWatch investigates a novel variant with its genomic context, analyzes the functional effect if it is located in a protein coding region or in a GT-AG splice site, presents information of genes nearby, checks affection to ESE and ESS hexamers pattern [from Rescue-ESE (17) and Fas-ESS (18)] if the variant is in an exon, and predicts risk of the variant based on the above-mentioned information. If the variant is reported in dbSNP or 1000 Genomes Project (19), related details will be listed as well.
Creating an annotation database for VarioWatch not only improves the system performance, but also enables VarioWatch to serve more than one reference version at the same time. VarioWatch currently provides annotations of two popular human genome reference versions (NCBI build 36.3, NCBI build 37.2), including gene annotation, pre-computed variation risks, known variants from dbSNP, 1000 Genomes Project (released on October 2011), OMIM (20) and other minor variant databases (see Supplementary Data).
INPUT
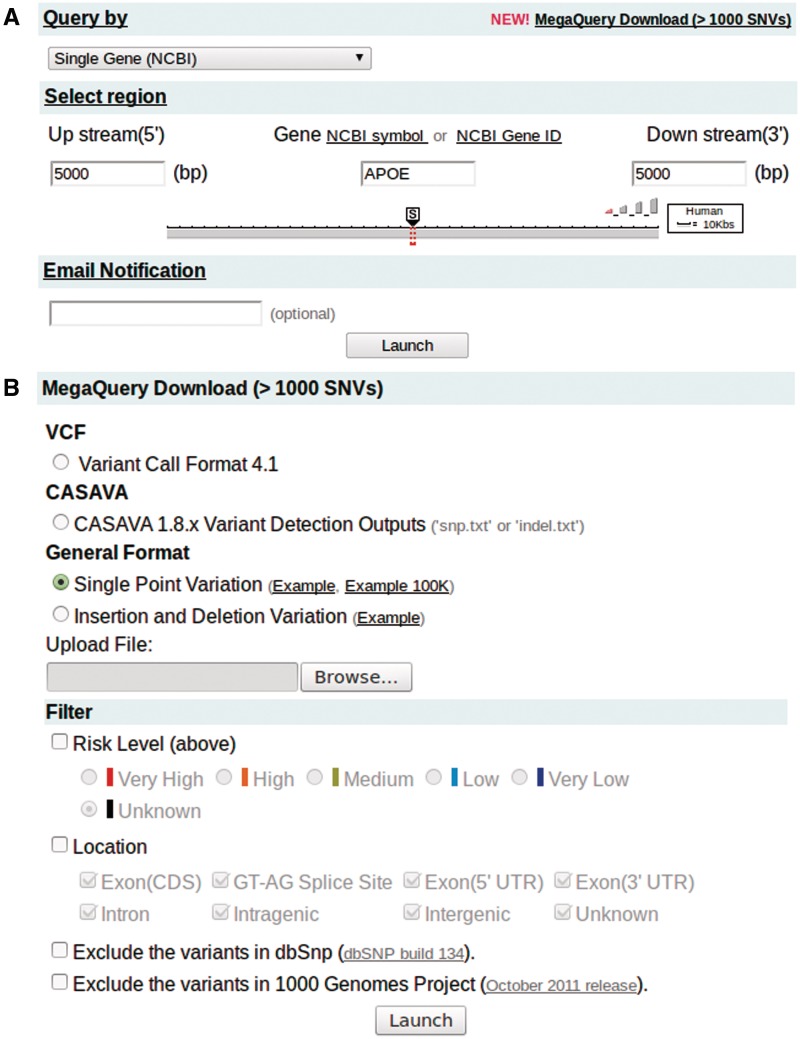
Users can easily query and visualize up to 1000 regions by chromosome positions, markers, gene symbols, a batch file input, etc. (Figure 1A). For instance, they can use a physical position, a single marker (e.g. SNP), plus downstream and upstream spans, to define a chromosome region like in GenoWatch. VarioWatch also supports sequence upload, finding all variants on the uploaded sequence by BLAT (21) and then annotating them automatically. By incorporating human variation data sets, such as OMIM, VarioWatch allows a disease name query. It first translates the input disease name into a group of relevant genes then shows all annotations of these genes as well as variants within.
Figure 1.
Input pages for normal query and MegaQuery. (A) An example to retrieve and visualize genomic annotations on gene APOE plus 5000 bases upstream and downstream. (B) MegaQuery Download is capable of taking a massive amount of variants as input, labeling them with genomic annotations, filtering out unwanted records and returning with purified annotation results.
Furthermore, VarioWatch has a special unit called MegaQuery (Figure 1B) dedicated to annotating millions of variants generated by NGS. MegaQuery currently supports batch queries for both single nucleotide substitution and in/del variants. Users can upload a file containing a list of variants. Examples are provided for different input types, respectively. Result files, e.g. snp.txt or indel.txt from Illumina CASAVA variant detection outcome or VCF format, can also be directly uploaded through MegaQuery to process.
Often, instead of examining all the variants identified by NGS, researchers only want to examine those which satisfy their research needs. Before, upon receiving variant annotation data, they either looked for further help from an IT specialist or turned to a computer-based spreadsheet, doing tedious work to achieve this goal. To address this issue, MegaQuery provides four handy filters to help researchers listing variants with functional impacts, with predicted risk above a certain threshold, in specific gene region or variants not reported in either dbSNP or 1000 Genome Project.
OUTPUT
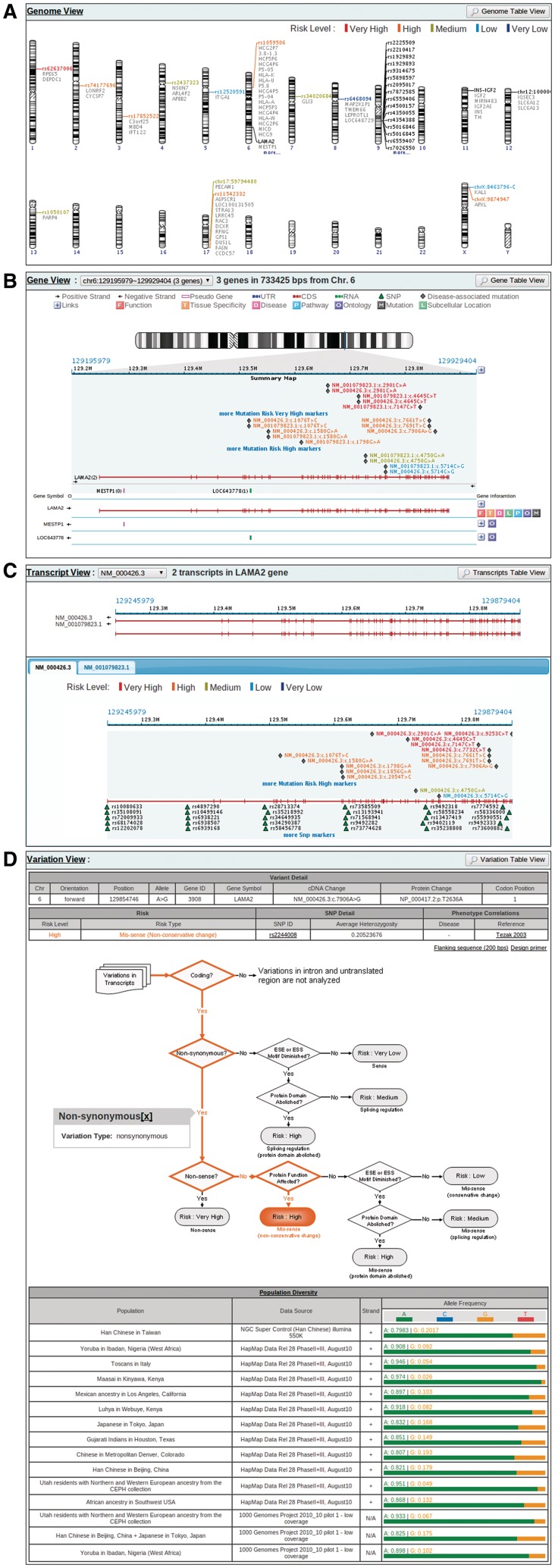
The results page is comprised of Genome View, Gene View, Transcript View and Variation View. Genome View and Gene View are generally inherited from GenoWatch. Genome View (Figure 2A) displays an overview of input markers plus their nearby genes. If a marker is a variant with risky functional impact, it is coloured according to the risk level. Clicking on a marker leads to Gene View (Figure 2B), showing structured genes and their corresponding annotations, including gene functions, tissue-specificity, diseases and so on. Instead of showing only SNP annotations like in GenoWatch, VarioWatch also lists disease-associated mutations and reveals the relation between query variants and these known mutations in this view. Transcript View (Figure 2C) presents transcript structure, the functional impacts of the same variant on different transcript isoforms and the distribution of known variants within. Variation View (Figure 2D) discloses the annotation details of a variant. It comprises three areas. The top area tabulates detailed variant information including its location, allele change, gene ID and gene symbol if the variant sits in a gene, cDNA change if the variant causes transcript change, protein and codon change if the variant falls in a translated region, estimated risk level, SNP information if the variant is a SNP, related disease and literature reference. The middle area graphs a risk-level decision tree and a highlighted path to show how the risk level of the variant is decided. Users can click on the path steps to obtain detailed reasons and references to data sources. What’s more, at the upper right corner of the area are links for users to download the variant-containing sequence and design primers for that variant. Finally, at the bottom area, information of population diversity extracted from 1000 Genomes project and HapMap (22) is clearly presented. All views can be exported to a text file for further analysis.
Figure 2.
Example output pages for visualized annotation result. (A) Genome View provides a bird's-eye-view of the query result on the genome scale. It shows the distribution of the query items on the whole genome, and colours each item according to the risk level analyzed based on the annotation results. (B) Gene View displays each query item in the context of genes and mutations known to cause diseases. In addition to providing a diagram representation of gene structures, including introns and exons, it also annotates each gene within the view-port with known functions, tissue specificity, ontology, pathway involved and disease caused. Disease-relevant mutations are also revealed. This view was designed with the aim to expedite gene-relevant literature searching. (C) Transcript View displays a query item in the transcript context. Since one variant may have different effects on different transcript isoforms, this view provides a precise genomic context in which the query item is analyzed. Transcript View also depicts known SNPs within the specified transcript along with disease-relevant mutations. (D) Variation View shows the annotation details of a query item, the decision tree of risk evaluation, and the relevant allele frequencies in different human races.
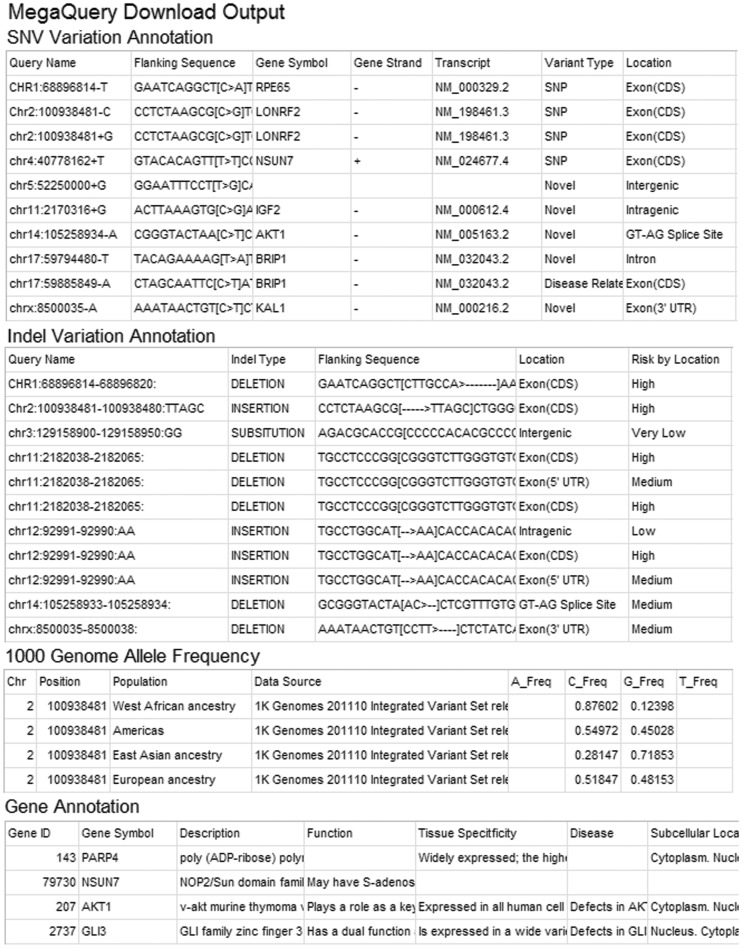
The results downloaded through MegaQuery are a zip file containing three reports: SNV/Indel Variation Annotation, 1000 Genome Allele Frequency and Gene Annotation (Figure 3). The three CSV-formatted reports have the same contents as a results page minus the visualization part and reference literature. Users can visualize any individual variant by clicking the URL provided in the last column of the SNV/Indel Variant Annotation report. Also, users can further manipulate these files with any application that supports CSV file format.
Figure 3.
MegaQuery Download responds a query with one zip file containing three different reports: SNV/Indel Variation Annotation, 1000 Genome Allele Frequency and Gene Annotation. SNV variation annotation provides a text-based annotation and risk analysis result of each query item in CSV format, while the other two auxiliary reports provide relevant allele frequencies and the information of containing genes.
IMPLEMENTATION
VarioWatch is written in Java programming language with Struts framework and JDBC technology. To further improve user experience, JavaScript is used for rendering the interactive input and output page. This makes it easier for users to define a genomic region in query page and to browse the classified result page.
For VarioWatch database construction, we built a script that mirrors all needed source data files from public domain FTP sites. Once each data source is verified to be consistent with their reference version, a pipe-line system will be involved to process these data into databases. In addition, a simple computer cluster system is built for hosting SIFT non-synonymous variants prediction tool (23). Combining these pre-computed and stored results, each variant generated from all possible substitution bases in coding regions and GT-AG splice sites is given a functional risk level and type.
CONCLUSION
VarioWatch provides an easy way for researchers to directly and quickly annotate a large number of human genomic variants online without having to run an offline annotating application or needing help from an IT specialist. The annotation is comprehensive. The input interface is intuitive and the returning outcome is displayed in a carefully designed results page. Its reliability, availability and serviceability are much better than GenoWatch because of database localization. VarioWatch should be able to help researchers facilitate their work substantially in variant annotation and prioritization in the NGS era.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table 1 and Supplementary References [24–26].
FUNDING
Academia Sinica Life Sciences [40-05-GMM]; National Science Council, Taiwan, R.O.C. [NSC100-2319-B-001-001]; National Center for Genomic Medicine. Funding for open access charge: Academia Sinica Life Sciences [40-05-GMM].
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
Special thanks to Dr Jer-Yuarn Wu, Director of National Center for Genome Medicine, Academia Sinica, for his support of this work and to Ms Stephanie Dee for editing the article.
REFERENCES
- 1.Wang J, Ronaghi M, Chong SS, Lee CG. pfSNP: an integrated potentially functional SNP resource that facilitates hypotheses generation through knowledge syntheses. Hum. Mutat. 2011;32:19–24. doi: 10.1002/humu.21331. [DOI] [PubMed] [Google Scholar]
- 2.Chelala C, Khan A, Lemoine NR. SNPnexus: a web database for functional annotation of newly discovered and public domain single nucleotide polymorphisms. Bioinformatics. 2009;25:655–661. doi: 10.1093/bioinformatics/btn653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schmitt AO, Assmus J, Bortfeldt RH, Brockmann GA. CandiSNPer: a web tool for the identification of candidate SNPs for causal variants. Bioinformatics. 2010;26:969–970. doi: 10.1093/bioinformatics/btq068. [DOI] [PubMed] [Google Scholar]
- 4.Riva A, Kohane IS. SNPper: retrieval and analysis of human SNPs. Bioinformatics. 2002;18:1681–1685. doi: 10.1093/bioinformatics/18.12.1681. [DOI] [PubMed] [Google Scholar]
- 5.DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ge D, Ruzzo EK, Shianna KV, He M, Pelak K, Heinzen EL, Need AC, Cirulli ET, Maia JM, Dickson SP, et al. SVA: software for annotating and visualizing sequenced human genomes. Bioinformatics (Oxford, England) 2011;27:1998–2000. doi: 10.1093/bioinformatics/btr317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Makarov V, O'Grady T, Cai G, Lihm J, Buxbaum JD, Yoon S. AnnTools: a comprehensive and versatile annotation toolkit for genomic variants. Bioinformatics (Oxford, England) 2012;28:724–725. doi: 10.1093/bioinformatics/bts032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen YH, Liu CK, Chang SC, Lin YJ, Tsai MF, Chen YT, Yao A. GenoWatch: a disease gene mining browser for association study. Nucleic Acids Res. 2008;36:W336–W340. doi: 10.1093/nar/gkn214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Benson DA, Karsch-Mizrachi I, Clark K, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Res. 2012;40:D48–D53. doi: 10.1093/nar/gkr1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pruitt KD, Tatusova T, Klimke W, Maglott DR. NCBI Reference Sequences: current status, policy and new initiatives. Nucleic Acids Res. 2009;37:D32–D36. doi: 10.1093/nar/gkn721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.UniProt Consortium. Reorganizing the protein space at the Universal Protein Resource (UniProt) Nucleic Acids Res. 2012;40:D71–D75. doi: 10.1093/nar/gkr981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32:D277–D280. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barrell D, Dimmer E, Huntley RP, Binns D, O'Donovan C, Apweiler R. The GOA database in 2009: an integrated Gene Ontology Annotation resource. Nucleic Acids Res. 2009;37:D396–D403. doi: 10.1093/nar/gkn803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu CK, Chen YH, Tang CY, Chang SC, Lin YJ, Tsai MF, Chen YT, Yao A. Functional analysis of novel SNPs and mutations in human and mouse genomes. BMC Bioinformatics. 2008;9(Suppl. 12):S10. doi: 10.1186/1471-2105-9-S12-S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fairbrother WG, Yeo GW, Yeh R, Goldstein P, Mawson M, Sharp PA, Burge CB. RESCUE-ESE identifies candidate exonic splicing enhancers in vertebrate exons. Nucleic Acids Res. 2004;32:W187–W190. doi: 10.1093/nar/gkh393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Z, Rolish ME, Yeo G, Tung V, Mawson M, Burge CB. Systematic identification and analysis of exonic splicing silencers. Cell. 2004;119:831–845. doi: 10.1016/j.cell.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 19.1000 Genomes Project Consortium. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Amberger J, Bocchini C, Hamosh A. A new face and new challenges for Online Mendelian Inheritance in Man (OMIM(R)) Hum. Mutat. 2011;32:564–567. doi: 10.1002/humu.21466. [DOI] [PubMed] [Google Scholar]
- 21.Kent WJ. BLAT: the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.International HapMap Consortium. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ng PC, Henikoff S. SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bruno AE, Li L, Kalabus JL, Pan Y, Yu A, Hu Z. miRdSNP: a database of disease-associated SNPs and microRNA target sites on 3′UTRs of human genes. BMC genomics. 2012;13:44. doi: 10.1186/1471-2164-13-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cariaso M, Lennon G. SNPedia: a wiki supporting personal genome annotation, interpretation and analysis. Nucleic Acids Res. 2012;40:D1308–D1312. doi: 10.1093/nar/gkr798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, Manolio TA. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl Acad. Sci. USA. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]