Abstract
Homozygosity mapping is a common method to map recessive traits in consanguineous families. To facilitate these analyses, we have developed HomozygosityMapper, a web-based approach to homozygosity mapping. HomozygosityMapper allows researchers to directly upload the genotype files produced by the major genotyping platforms as well as deep sequencing data. It detects stretches of homozygosity shared by the affected individuals and displays them graphically. Users can interactively inspect the underlying genotypes, manually refine these regions and eventually submit them to our candidate gene search engine GeneDistiller to identify the most promising candidate genes. Here, we present the new version of HomozygosityMapper. The most striking new feature is the support of Next Generation Sequencing *.vcf files as input. Upon users’ requests, we have implemented the analysis of common experimental rodents as well as of important farm animals. Furthermore, we have extended the options for single families and loss of heterozygosity studies. Another new feature is the export of *.bed files for targeted enrichment of the potential disease regions for deep sequencing strategies. HomozygosityMapper also generates files for conventional linkage analyses which are already restricted to the possible disease regions, hence superseding CPU-intensive genome-wide analyses. HomozygosityMapper is freely available at http://www.homozygositymapper.org/.
INTRODUCTION
Linkage analysis is still widely considered the ‘gold standard’ for disease gene mapping. However, especially in complex consanguineous families, these analyses require high-performance computers. Even then, a multi-point analysis of a medium-sized genotyping array with 100 000 single nucleotide polymorphisms (SNPs) may take weeks. In one of our benchmarking experiments, the analysis of 50 000 markers in a single consanguineous family needed more than 12 weeks to complete. Although it is possible to employ only a subset of a few thousand markers in the initial analysis and to re-analyse only the homozygous regions with the complete marker set, such an analysis can still take several hours. To overcome the restraints posed by linkage software, we have developed HomozygosityMapper (1), a web-based approach to homozygosity mapping.
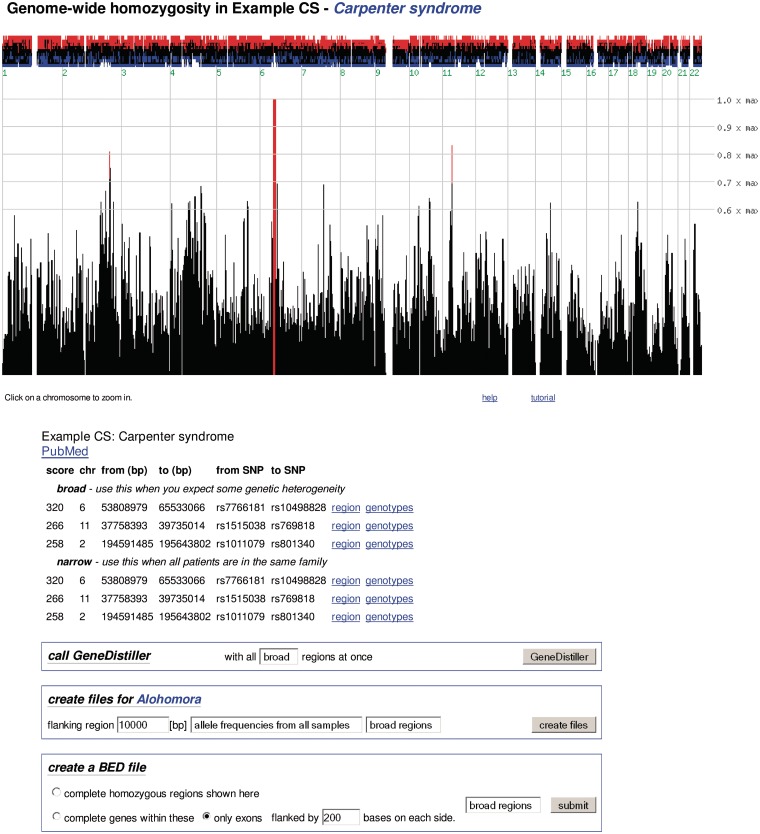
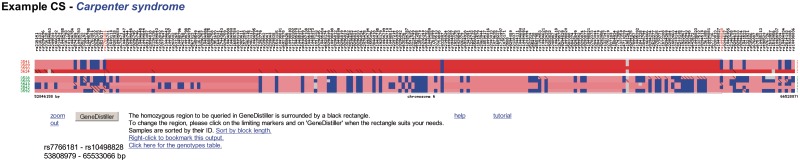
As the basic concept of homozygosity mapping is to trace the inheritance of the same chromosomal region from an ancestor via two consanguineous heterozygous parents and hence homozygosity in the patients, the disease region must be homozygous in all affected family members. It is thus not necessary to waste CPU time on a lengthy whole genome multipoint linkage analysis only to search for homozygous regions in the patients. Several applications (1–5), including HomozygosityMapper, therefore simply detect homozygous stretches in the patients and score them according to their length. In contrast to the other tools, HomozygosityMapper is entirely web-based so that no software installation is required. Users can upload their genotype files into our database without the need for reformatting, define the samples that represent affected or healthy individuals and immediately start the search for homozygosity. Further information such as marker positions or allele frequencies is already stored in the database. After the analysis, the genome-wide homozygosity is plotted against the marker coordinates, interesting regions are highlighted in the plot and listed in a table (Figure 1). The application also offers the users the ability to inspect single chromosomes. Furthermore, the underlying genotypes can be displayed in a colour-coded matrix plot that highlights long homozygous regions (Figure 2). The entire process of upload, analysis and display is completed within 5 min for a 50 K genotyping project with six samples; arrays featuring one million SNPs are completed in less than 30 min.
Figure 1.
Genome-wide homozygosity. This screen shot shows the genome-wide homozygosity scores produced by HomozygosityMapper. These are plotted as a bar chart with red bars indicating the most promising genomic regions. Clicking on a bar will zoom into the chromosome. Above the bar chart, the excess or shortage of homozygous genotypes in cases versus controls is depicted. Below the figure, direct links to the most interesting regions are given and data export possibilities are provided. All figures depict the Carpenter syndrome study (6).
Figure 2.
Genotypes view. HomozygosityMapper also displays the genotypes of all samples. Here, the markers are placed on the x-axis while the samples are on the y-axis, with the patients on top and with red IDs. Genotypes are colour-coded: grey, unknown, blue, heterozygous, red, homozygous stretches (colour saturation reflects the length of the stretch). This figure also reveals the presence of a single heterozygous marker within the homozygous region (possibly a genotyping error and ignored by HomozygosityMapper). The patient on the bottom is from another family than the first two and does not share the same haplotype over the entire homozygous stretch. This can be seen from the genotypes with the diagonal bar indicating the less abundant of the homozygous genotypes. Users are free to change the boundaries of the region and can subsequently submit this region to GeneDistiller.
HomozygosityMapper provides various links to our candidate gene search engine GeneDistiller (7). It has hence become very convenient to proceed from the genotype file to the search for candidate genes. The process is so simple and intuitive that clinicians or researchers are able to analyse their data on their own without the need to consult dedicated IT specialists.
The advent of Next Generation Sequencing (NGS) approaches has recently shifted the strategy to identify the disease mutation from sequencing candidate genes (8) to sequencing entire chromosomal regions (9). We expect that due to the falling costs, NGS will increasingly be applied no longer only for mutation detection after a linkage analysis but indeed as a single method to combine both (10).
Here, we present the novel version of HomozygosityMapper that generates the *.bed files needed to include all possible disease regions in targeted enrichment deep sequencing. The software also covers the opposite approach, i.e. a fast homozygosity mapping on whole genome or whole exome sequencing data produced before a linkage analysis. According to our users’ wishes, we have extended the software to handle other species than humans (11). Our database currently includes the most common model organisms and farm animals but can be extended to other species in short time. Furthermore, we have refined the options to handle single families and loss of heterozygosity studies.
A detailed description of the approach, the features of HomozygosityMapper and data on its performance is provided in the original publication (1) and on our website.
CHANGES IN THE NEW VERSION
Since its introduction, HomozygosityMapper has become a widely used tool for homozygosity mapping (8,9,11–15). By winter 2011–2012, 8 billion genotypes, created in more than 3300 mapping projects, have been permanently stored in our database. As users are free to use the software without registration and to delete their data after the analysis, the actual number of analysed projects is probably even higher. Given the large number of users, we received numerous suggestions how to improve our software. These ranged from bug reports to completely new features. In the new version of HomozygosityMapper, we have integrated many of these.
Import of NGS genotypes
The most striking new feature of HomozygosityMapper is the integration of NGS data. Users can now directly upload the Variant Call Format (*.vcf) files generated in NGS projects. HomozygosityMapper will then pick all positions in which either variations from the RefSeq are found or which are known to bear SNPs and store the genotypes. We provide a description of the import file format and the generation of these files with SAMtools (16) on our website.
Integration of additional species
As homozygosity mapping is also employed by animal breeders and by researchers working on model organisms, we had several requests for animal versions. An early prototype was successfully employed in the mapping of generalised progressive retinal atrophy in dogs (11). We have now integrated a framework to include an unlimited number of species into the application on short notice. So far, data for seven different model organisms or breeding animals (humans, cattle, dogs, horses, mice, rats and sheep) are stored in our database. We encourage our users to contact us requesting the integration of other species.
Optimisation of single family approaches
The original release of HomozygosityMapper was aimed at the classic setting for homozygosity mapping with cases from different consanguineous families where genetic homogeneity of the disease was not absolutely sure. In single families, however, affected individuals should carry the same disease haplotype. We have now included an option to require genetic homogeneity around the disease locus.
Users can also decide to include the genetic information of healthy siblings to further narrow down the disease region considerably. In contrast to the standard settings, this approach does not only search for regions that are homozygous in many affected individuals but also for a homozygous disease haplotype shared by all affected individuals which must not be present in any of the healthy controls.
Optimisation of loss of heterozygosity studies and genetic heterogeneity
Studies for loss of heterozygosity (or microdeletions) are often performed on large cohorts of unrelated subjects with similar phenotypes. Here, a high degree of heterogeneity is possible (17,18). We have therefore added an option to exclude very short homozygous regions because these may occur due to uninformative makers and also by chance. In this mode, the search focuses on long regions only shared by some patients. This approach is also useful when genetic heterogeneity is expected in a classic homozygosity mapping setting, e.g. with patients from different regions or with slightly different phenotypes.
Improved transition to GeneDistiller
We have optimised the transition between the identification of homozygous stretches in the genome and the analysis of potential candidate genes therein. Users can now seamlessly switch to the candidate gene search at any step after the analysis. They can search for homozygosity around candidate genes, query the genes contained in all interesting regions at once or view the genotypes within a single region, refine the region and search among the genes it contains.
File export for linkage analysis
As homozygosity is the a priori condition for homozygosity mapping, it is not necessary to perform a CPU-intensive whole genome linkage analysis when searching for homozygosity in consanguineous families. The search for linkage can be restricted to longer stretches of homozygosity shared by all or at least many patients. HomozygosityMapper now offers the export of genotype and map files for the potential disease regions in the format used by ALOHOMORA (19). ALOHOMORA is a tool that converts SNP genotypes and marker information into the input files required by common linkage analysis software. Using this approach, a multipoint linkage analysis can be restricted to the possible disease regions thus sparing considerable CPU time.
File export for NGS
With the advent of NGS technologies, it has become feasible to search for mutations in numerous genes or even in complete linkage intervals in one run. In the case of homozygosity mapping, all genes that are positional candidates, i.e. located in homozygous regions, thus can be sequenced simultaneously. HomozygosityMapper can now generate the *.bed files needed for the targeted DNA capture of the possible disease regions. The files can either cover (a) all homozygous regions completely, (b) only the genes contained within them or (c) only the exons plus a user-defined flanking region.
Performance
Because of the increasing use of HomozygosityMapper with very large datasets (the most prominent were 200 million genotypes from 1000 samples genotyped with Affymetrix Axiom chips), we have restructured our database for a better performance on huge datasets, mainly by adding further indices, a different commit strategy and an adapted configuration. We have recently acquired a new RAID system that will further increase query speed.
Privacy
The original version of HomozygosityMapper required users to login to create projects only visible to themselves. We have now added the possibility to make a project private without a user account. In these cases, a secret key is issued when the genotypes are uploaded. Such projects can only been accessed with this key and they are not displayed in any lists. However, in these cases, users lose access to their data if they lose their key. Of course, this key can be shared with collaborators but this will grant them unlimited access to the data.
New data import formats
Besides the integration of *.vcf files, we have extended the possible genotyping arrays to allow the import of genotypes generated on recent arrays such as the Affymetix Axiom family. We have also adapted the import routine so that further file formats (some of which were in-house formats of other groups) are accepted. We will gladly add further possible arrays and formats on request.
Implementation
HomozygosityMapper was programmed in Perl. It makes use of a PostgreSQL 8.3 database. Web server and database run on an Intel Xeon platform with two QuadCore processors and 48 GB of RAM under Fedora Core Linux. A thorough description of the implementation can be found on the website.
The website was developed with and optimised for Mozilla Firefox 2-10. It was successfully tested with Firefox 2-10 (under different versions of Linux, Microsoft Windows and MacOS) and Microsoft Internet Explorer 6, 7 and 8.
Future plans
We are permanently improving and extending HomozygosityMapper. The next milestone will be the tight integration of MutationTaster (20), our web-based tool to predict the disease potential of DNA alterations, into the analysis of deep sequencing genotypes. With a future interface, users will be able to retrieve a list of all homozygous variants that are located in possible disease regions and have a high disease potential. We will add new species, new genotyping assays and support for new file formats upon request on short notice.
CONCLUSION
We have presented the novel version of HomozygosityMapper, a web-based application aimed at homozygosity mapping of SNP genotypes and NGS data in different species. HomozygosityMapper is freely accessible at http://www.homozygositymapper.org/ and there is no login requirement. We provide a step-by-step tutorial and a detailed documentation on our website.
FUNDING
Funding for open access charge: Deutsche Forschungsgemeinschaft (DFG) via the NeuroCure Cluster of Excellence [Exc 257] at the Charité, Berlin; Einstein Foundation, Berlin [A-2011-63].
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank Regina Kropatsch for beta-testing the dog version and Evelyn Lüdeking for the critical review of the manuscript. They also thank the numerous users who provided bug reports and suggested new features.
REFERENCES
- 1.Seelow D, Schuelke M, Hildebrandt F, Nürnberg P. HomozygosityMapper—an interactive approach to homozygosity mapping. Nucleic Acids Res. 2009;37:W593–W599. doi: 10.1093/nar/gkp369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Woods CG, Valente EM, Bond J, Roberts E. A new method for autozygosity mapping using single nucleotide polymorphisms (SNPs) and EXCLUDEAR. J. Med. Genet. 2004;41:e101. doi: 10.1136/jmg.2003.016873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carr IM, Flintoff KJ, Taylor GR, Markham AF, Bonthron DT. Interactive visual analysis of SNP data for rapid autozygosity mapping in consanguineous families. Hum. Mutat. 2006;27:1041–1046. doi: 10.1002/humu.20383. [DOI] [PubMed] [Google Scholar]
- 4.Carr IM, Szymanska K, Sheridan E, Markham AF, Bonthron DT, Johnson CA. Shadow autozygosity mapping by linkage exclusion (SAMPLE): a simple strategy to identify the genetic basis of lethal autosomal recessive disorders. Hum. Mutat. 2009;30:1642–1649. doi: 10.1002/humu.21105. [DOI] [PubMed] [Google Scholar]
- 5.Carr IM, Sheridan E, Hayward BE, Markham AF, Bonthron DT. IBDfinder and SNPsetter: tools for pedigree-independent identification of autozygous regions in individuals with recessive inherited disease. Hum. Mutat. 2009;30:960–967. doi: 10.1002/humu.20974. [DOI] [PubMed] [Google Scholar]
- 6.Jenkins D, Seelow D, Jehee FS, Perlyn CA, Alonso LG, Bueno DF, Donnai D, Josifiova D, Mathijssen IMJ, Morton JEV, et al. RAB23 Mutations in Carpenter Syndrome Imply an Unexpected Role for Hedgehog Signaling in Cranial-Suture Development and Obesity. Am. J. Hum. Genet. 2007;80:1162–1170. doi: 10.1086/518047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seelow D, Schwarz JM, Schuelke M. GeneDistiller—distilling candidate genes from linkage intervals. PLoS One. 2008;3:e3874. doi: 10.1371/journal.pone.0003874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zelinger L, Banin E, Obolensky A, Mizrahi-Meissonnier L, Beryozkin A, Bandah-Rozenfeld D, Frenkel S, Ben-Yosef T, Merin S, Schwartz SB, et al. A missense mutation in DHDDS, encoding dehydrodolichyl diphosphate synthase, is associated with autosomal-recessive retinitis pigmentosa in Ashkenazi Jews. Am. J. Hum. Genet. 2011;88:207–215. doi: 10.1016/j.ajhg.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bolze A, Byun M, McDonald D, Morgan NV, Abhyankar A, Premkumar L, Puel A, Bacon CM, Rieux-Laucat F, Pang K, et al. Whole-exome-sequencing-based discovery of human FADD deficiency. Am. J. Hum. Genet. 2010;87:873–881. doi: 10.1016/j.ajhg.2010.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Biesecker LG. Exome sequencing makes medical genomics a reality. Nat. Genet. 2010;42:13–14. doi: 10.1038/ng0110-13. [DOI] [PubMed] [Google Scholar]
- 11.Kropatsch R, Petrasch-Parwez E, Seelow D, Schlichting A, Gerding WM, Akkad DA, Epplen JT, Dekomien G. Generalized progressive retinal atrophy in the Irish Glen of Imaal Terrier is associated with a deletion in the ADAM9 gene. Mol. Cell. Probes. 2010;24:357–363. doi: 10.1016/j.mcp.2010.07.007. [DOI] [PubMed] [Google Scholar]
- 12.Rafiq MA, Ansar M, Marshall CR, Noor A, Shaheen N, Mowjoodi A, Khan MA, Ali G, Amin-ud-Din M, Feuk L, et al. Mapping of three novel loci for non-syndromic autosomal recessive mental retardation (NS-ARMR) in consanguineous families from Pakistan. Clin. Genet. 2010;78: 478–483. doi: 10.1111/j.1399-0004.2010.01405.x. [DOI] [PubMed] [Google Scholar]
- 13.Rajab A, Straub V, McCann LJ, Seelow D, Varon R, Barresi R, Schulze A, Lucke B, Lützkendorf S, Karbasiyan M, et al. Fatal cardiac arrhythmia and long-QT syndrome in a new form of congenital generalized lipodystrophy with muscle rippling (CGL4) due to PTRF-CAVIN mutations. PLoS Genet. 2010;6:e1000874. doi: 10.1371/journal.pgen.1000874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boyden SE, Salih MA, Duncan AR, White AJ, Estrella EA, Burgess SL, Seidahmed MZ, Al-Jarallah AS, Alkhalidi HMS, Al-Maneea WM, et al. Efficient identification of novel mutations in patients with limb girdle muscular dystrophy. Neurogenetics. 2010;11:449–455. doi: 10.1007/s10048-010-0250-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Birk E, Har-Zahav A, Manzini CM, Pasmanik-Chor M, Kornreich L, Walsh CA, Noben-Trauth K, Albin A, Simon AJ, Colleaux L, et al. SOBP is mutated in syndromic and nonsyndromic intellectual disability and is highly expressed in the brain limbic system. Am. J. Hum. Genet. 2010;87:694–700. doi: 10.1016/j.ajhg.2010.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Helbig I, Mefford HC, Sharp AJ, Guipponi M, Fichera M, Franke A, Muhle H, de Kovel C, Baker C, von Spiczak S, et al. 15q13.3 Microdeletions increase risk of idiopathic generalized epilepsy. Nat. Genet. 2009;41:160–162. doi: 10.1038/ng.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maleno I, Aptsiauri N, Cabrera T, Gallego A, Paschen A, López-Nevot MA, Garrido F. Frequent loss of heterozygosity in the β2-microglobulin region of chromosome 15 in primary human tumors. Immunogenetics. 2011;63:65–71. doi: 10.1007/s00251-010-0494-4. [DOI] [PubMed] [Google Scholar]
- 19.Rüschendorf F, Nürnberg P. ALOHOMORA: a tool for linkage analysis using 10K SNP array data. Bioinformatics. 2005;21:2123–2125. doi: 10.1093/bioinformatics/bti264. [DOI] [PubMed] [Google Scholar]
- 20.Schwarz JM, Rödelsperger C, Schuelke M, Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat. Methods. 2010;7:575–576. doi: 10.1038/nmeth0810-575. [DOI] [PubMed] [Google Scholar]