Abstract
The many ongoing genome sequencing initiatives are delivering comprehensive lists of the individual molecular components present in an organism, but these reveal little about how they work together. Follow-up initiatives are revealing thousands of interrelationships between gene products that need to be analyzed with novel bioinformatics approaches able to capture their complex emerging properties. Recently, we developed NetAligner, a novel network alignment tool that allows the identification of conserved protein complexes and pathways across organisms, providing valuable hints as to how those interaction networks evolved. NetAligner includes the prediction of likely conserved interactions, based on evolutionary distances, to counter the high number of missing interactions in current interactome networks, and a fast assessment of the statistical significance of individual alignment solutions, which increases its performance with respect to existing tools. The web server implementation of the NetAligner algorithm presented here features complex, pathway and interactome to interactome alignments for seven model organisms, namely Homo sapiens, Mus musculus, Drosophila melanogaster, Caenorhabditis elegans, Arabidopsis thaliana, Saccharomyces cerevisiae and Escherichia coli. The user can query complexes and pathways of arbitrary topology against a target species interactome, or directly compare two complete interactomes to identify conserved complexes and subnetworks. Alignment solutions can be downloaded or directly visualized in the browser. The NetAligner web server is publicly available at http://netaligner.irbbarcelona.org/.
INTRODUCTION
Genome sequencing projects are providing nearly complete repertoires of the genes and proteins present in many species. However, most biological systems are highly complex, and knowing the individual components cannot explain the function of biological processes, which are orchestrated through intricate networks of protein–protein interactions. Accordingly, many efforts were directed toward uncovering the hundreds of thousands of interrelationships between macromolecules, creating the first interactome drafts for several important model organisms, including human (1–3).
Interpreting this huge amount of data to discover emerging principles and study the evolution of protein complexes and pathways demands novel bioinformatics tools. Similar to the importance of sequence alignment for studying genome function and evolution, network alignment is expected to be crucial for understanding the global organization of whole species interactomes and for discovering how specific protein interaction networks have evolved (4,5).
The tens, or even hundreds, of thousands of interactions within an organism and the complex homology relationships between different species represent a difficult challenge for network alignment methods, and particularly for web server implementations with restraints on computational resources. The few existing tools are thus limited to one specific task, either the alignment of linear pathways (6), the querying of well-connected complexes (7) or the detection of conserved functional modules across species (8–10). Other limitations are related to the difficulty in handling the large fraction of false positives and negatives present in the current versions of interactome networks (11). To address these challenges, we recently developed the NetAligner algorithm, a novel network alignment method that considers interaction reliabilities and protein similarities to detect potential false positives, as well as the prediction of likely conserved interactions, based on evolutionary distances, to counter the high number of missing interactions in current interactome networks (12). In addition, NetAligner allows gaps and mismatches in the alignment to account for small amounts of network rewiring during evolution and employs a fast Monte-Carlo permutation test to assess the statistical significance of alignment solutions. All these resulted in an accurate and efficient implementation of the NetAligner search strategy that allows not only the alignment of large protein complexes and pathways of arbitrary topology to whole species interactomes but also full interactome to interactome alignments.
To make NetAligner accessible to the biological community in a fully integrated and easy to use fashion, we have implemented a web server that retains almost the entire functionality of the stand-alone application and where the data for the seven most commonly used model organisms (i.e. Homo sapiens, Mus musculus, Drosophila melanogaster, Caenorhabditis elegans, Arabidopsis thaliana, Saccharomyces cerevisiae and Escherichia coli) is already included. The web server is publicly available at http://netaligner.irbbarcelona.org/.
THE NetAligner WEB SERVER
Alignment tasks
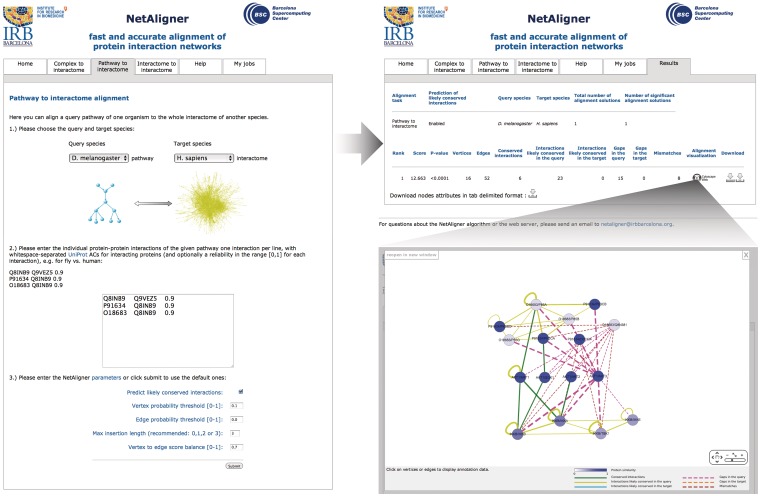
Our web server implementation of the NetAligner algorithm (12) features three different alignment tasks: ‘Complex to interactome’, ‘Pathway to interactome’ and ‘Interactome to interactome’ alignments. Each of these tasks can be selected by clicking on the respective tab (Figure 1). On the ‘Complex to interactome’ page, the user can align a query complex of one species to the whole interactome of either the same or other species, to search for alternative complexes that might fulfill the same or a similar function within the cell and to identify conserved protein complexes across different organisms, respectively. The protein components of the given query complex have to be provided as a simple list of UniProt Accession Codes (ACs) (13). We benchmarked this task on a set of 71 conserved human/yeast complex pairs, from the manually curated MPACT (14) and CORUM (15) databases, and found that NetAligner could correctly identify as the top-ranked significant solution 55% of the query complexes with a precision of 81%, on average [see Ref. 12 for details]. Analogously, on the ‘Pathway to interactome’ page, a query pathway of one species can be aligned to the whole interactome of either the same or another species, to look for alternative signaling routes, backup circuits or pathway cross-talk, and to detect conserved pathways across organisms, respectively. Here, the list of protein–protein interactions of the given query pathway has to be supplied [in the form of whitespace-separated UniProt ACs (13)], and optionally a reliability estimate for each interaction (Figure 1). The performance of the method in this particular task was evaluated on a set of 19 human/fly, 32 human/yeast and 13 fly/yeast conserved pathway pairs originating from the KEGG database (16). We found that NetAligner correctly identified a significant solution for 55% of the query pathways with a precision of 80%, obtaining very similar results as for the complex to interactome alignment task, despite the much higher variation in the topologies of pathways [see Ref. 12 for details]. Finally, on the ‘Interactome to interactome’ page, it is possible to align the whole interactomes of two species to search for conserved protein complexes or subnetworks. In this case, NetAligner was able to automatically re-discover 44% of the known complexes common to human and yeast. Further details on the benchmark of the method can be found in (12), in which we show that NetAligner significantly outperforms both the current standard in the field for complex/pathway to interactome alignment, IsoRank (17), and the current standard for interactome to interactome alignment, NetworkBLAST (18), while requiring only a fraction of the runtime. Moreover, in contrast to other existing tools for network alignment (6,7,9,10,18), NetAligner is the first to allow both complex/pathway to interactome alignment and interactome to interactome alignment for networks of arbitrary topology. In addition, NetAligner also represents the first network alignment web server that offers the prediction of likely conserved interactions, based on evolutionary considerations, to counter the large number of false negatives in current interactome networks (11), significantly increasing alignment performance in several cases (12).
Figure 1.
Performing and visualizing network alignments with the NetAligner webserver. The NetAligner webserver can be used to perform ‘Complex to interactome’, ‘Pathway to interactome’ (shown on the left) and ‘Interactome to interactome’ alignments. After choosing one of the alignment tasks, the user can select the query and target species and enter a list of protein components or interactions for complex and pathway to interactome alignment, respectively, as well as the NetAligner parameters. Clicking the ‘Submit’ button will run NetAligner (12), and the ranked list of alignment results will be shown in a new tab once they are ready (top right). Clicking on the ‘Cytoscape Web’ (22) button next to a given alignment solution will display its network visualization (requires Flash support to be enabled in the browser), with vertex probabilities indicated by different shades of blue, ranging from 0 (white) to 1 (blue). Conserved interactions are shown in green and likely conserved interactions in the query and target species in yellow and light blue, respectively. Dashed lines highlight gaps in the query (magenta) and target network (orange), as well as mismatches (red).
NetAligner parameters
In all three alignment tasks, the user has to choose the query and target species, as well as the NetAligner parameters (Figure 1). These comprise the vertex and edge probability thresholds, which control the exclusion of vertices and edges with low probabilities, the max insertion length, which dictates the maximum number of protein insertions allowed in gaps and mismatches, as well as the vertex to edge score balance, which determines the impact of vertex and edge scores on the final score of alignment solutions [see the web server Help page and Ref. 12 for details]. Vertex and edge probability thresholds can be used to filter out distant homologs with only little sequence similarity and interactions with only low reliability or only low probability of being conserved, respectively, thus addressing the issue of false positive matchings. On the other hand, controlling the maximum insertion length of gaps and mismatches determines the amount of network rewiring during evolution considered in the alignment procedure. Although the maximum insertion length can be set arbitrarily high, due to the small-world property of current interactome networks (meaning that the average path length between any two proteins is small compared to their size) (19), we suggest not to set this parameter higher than three. In addition, the user can select the option to predict likely conserved interactions, based on evolutionary considerations [see Help page and Ref. 12 for details] to counter the prevalence of false negatives in current interactome networks (11). Although all parameters can be fine-tuned by the user, as a point of reference, we determined the optimal parameters for each task on the benchmark sets described above, which are displayed as default values in the web application.
The NetAligner search strategy
After selecting the NetAligner parameters, as well as the query and target species, and providing the list of complex components or protein interactions (for complex and pathway to interactome alignment, respectively), clicking the ‘Submit’ button (Figure 1) launches a NetAligner job on our server. In the case of complex to interactome alignment, since NetAligner expects two input networks, the server creates an induced complex network from the given list of protein components, using interaction data from the respective species interactome. In brief, NetAligner first collects all pairs of homologous proteins between the two input networks. Each of those pairs is represented by a vertex, except those with a probability below the given threshold. Vertex probabilities are calculated as the posterior probability of the respective proteins being real homologs given their BLAST E-value (20). The initial alignment graph is then constructed by drawing edges between vertices that are involved in a conserved or likely conserved interaction (if the option to predict likely conserved interactions was selected). Edges with a probability below the given threshold are filtered out, calculating edge probabilities based on the reliability estimates and conservation probabilities of the respective interactions. Alignment seeds are identified by searching for connected components in the initial alignment graph. That graph is then extended by connecting vertices of different seeds through gap or mismatch edges if the given homologs are connected by indirect interactions in one or both input networks, respectively. Again, the edge probability threshold is used to filter out false positives. Searching for connected components in the extended alignment graph then yields the final alignment solutions. Those are scored and ranked, and their statistical significance is determined using a Monte-Carlo permutation test. The score of each alignment solution is computed as the weighted sum over all vertex and edge scores, employing the user-defined vertex to edge score balance. Individual vertex and edge scores are calculated as the logarithm of the respective vertex or edge probability. For details about the NetAligner algorithm, please refer to the Help page and Ref. (12).
NetAligner output and visualization
A typical complex or pathway to interactome alignment query is computed in less than a minute, while interactome to interactome alignments usually take between 2 and 5 min, depending on the given species pair and NetAligner parameters. Once NetAligner has finished, the alignment results summary is displayed on a separate ‘Results’ tab, showing the type of alignment task performed, whether likely conserved interactions were predicted, the query and target species, as well as the total number of alignment solutions found and the number of statistically significant solutions. This is followed by the ranked list of all alignment solutions and their basic statistics, such as score, P-value and number of vertices and edges (Figure 1). Note that statistically insignificant solutions (marked in gray) might still be biologically relevant, due to underestimated interaction reliabilities or the incomplete nature of current interactomes (11). The URL of the results page, which can be bookmarked for future accession, contains the ID of the respective alignment job and is available on our server for 48 h. In addition, on the ‘My jobs’ page, the user can easily monitor the progress of different alignment jobs and access all last queries that he or she submitted. Each alignment solution can be downloaded as a simple tab-delimited file, in a standard format (the eXtensible Graph Markup and Modelling Language, XGMML), recognized by network visualization software like Cytoscape (21) or directly displayed in the browser by clicking on the ‘Cytoscape Web’ (22) button next to it (Figure 1). This Flash-based interactive visualization allows the user to select individual vertices and edges to get annotation data about the aligned proteins and types of interactions between them, as well as to navigate the alignment using the control panel (Figure 1).
DATA SOURCES
Collection of protein sequences
We collected protein sequences for human (H. sapiens), mouse (M. musculus), fly (D. melanogaster), worm (C. elegans), cress (A. thaliana), yeast (S. cerevisiae) and E. coli from the UniProt database release 2010_08 (13), including splice variants, but excluding cDNAs and fragments, as well as requiring experimental evidence on protein or transcript level. After clustering using UniRef 100 (13), we obtained non-redundant sets of 44 319 human, 42 706 mouse, 20 070 fly, 18 752 worm, 27 728 cress, 6203 yeast and 2726 E. coli protein sequences.
Whole organism interactomes
We constructed whole species interactomes by collecting all experimentally determined interactions from the public databases IntAct (23), MINT (24), HPRD (25), BioGRID (26), MatrixDB (27), MPIDB (28), InnateDB (29) and DIP (30). To transform protein complexes into binary interactions, we used the ‘spoke’ model whenever the bait of the given affinity purification was specified, and the ‘matrix’ model otherwise (31). We again applied the UniRef 100 (13) clustering to remove redundancy. Interactions involving proteins that were not in the respective species proteome were discarded, as well as all those interactions that could not be traced back to a publication or that were marked as ‘weak’ by the authors. We assigned reliabilities to each interaction based on the number of publications supporting it as defined by Kelley et al. (32). This resulted in non-redundant interactomes of 83 122 interactions in human, 14 853 in mouse, 30 168 in fly, 9471 in worm, 7921 in cress, 189 467 in yeast and 49 015 in E. coli.
Lists of homologous proteins
We determined lists of homologous proteins for all species combinations using proteome-wide all against all reciprocal BLAST (20) searches. To remove spurious hits, we required an E-value < 10−10 and considered only hits in the top ten of the BLASTP output.
All the data used by the web server is updated regularly twice per year, or whenever a new high-throughput interaction discovery experiment is released.
FUNDING
The Spanish Ministerio de Ciencia e Innovación [PSE-010000-2009-1 and BIO2010-22073, in part] and the European Commission under FP7 [Grant Agreement 223101 (AntiPathoGN)]. R.A.P. and A.C. are recipients of the Spanish Formación del Profesorado Universitario (FPU) and Juan de la Cierva fellowships, respectively. Funding for open access charge: BIO2010-22073.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We would like to thank Amelie Stein, Roberto Mosca, Andreas Zanzoni and Manuel Alonso (IRB Barcelona) for helpful discussions and for testing the web server.
REFERENCES
- 1.Rual J-F, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, Berriz GF, Gibbons FD, Dreze M, Ayivi-Guedehoussou N, et al. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- 2.Stelzl U, Worm U, Lalowski M, Haenig C, Brembeck FH, Goehler H, Stroedicke M, Zenkner M, Schoenherr A, Koeppen S, et al. A human protein-protein interaction network: a resource for annotating the proteome. Cell. 2005;122:957–968. doi: 10.1016/j.cell.2005.08.029. [DOI] [PubMed] [Google Scholar]
- 3.Ewing RM, Chu P, Elisma F, Li H, Taylor P, Climie S, McBroom-Cerajewski L, Robinson MD, O'Connor L, Li M, et al. Large-scale mapping of human protein-protein interactions by mass spectrometry. Mol. Syst. Biol. 2007;3:89. doi: 10.1038/msb4100134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kiemer L, Cesareni G. Comparative interactomics: comparing apples and pears? Trends Biotechnol. 2007;25:448–454. doi: 10.1016/j.tibtech.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 5.Beltrao P, Serrano L. Specificity and evolvability in eukaryotic protein interaction networks. PLoS Comput. Biol. 2007;3:e25. doi: 10.1371/journal.pcbi.0030025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kelley BP, Yuan B, Lewitter F, Sharan R, Stockwell BR, Ideker T. PathBLAST: a tool for alignment of protein interaction networks. Nucleic Acids Res. 2004;32:W83–W88. doi: 10.1093/nar/gkh411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bruckner S, Hüffner F, Karp RM, Shamir R, Sharan R. Topology-free querying of protein interaction networks. J. Comput. Biol. 2010;17:237–252. doi: 10.1089/cmb.2009.0170. [DOI] [PubMed] [Google Scholar]
- 8.Sharan R, Suthram S, Kelley RM, Kuhn T, McCuine S, Uetz P, Sittler T, Karp RM, Ideker T. Conserved patterns of protein interaction in multiple species. Proc. Natl Acad. Sci. USA. 2005;102:1974–1979. doi: 10.1073/pnas.0409522102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flannick J, Novak A, Srinivasan BS, McAdams HH, Batzoglou S. Graemlin: general and robust alignment of multiple large interaction networks. Genome Res. 2006;16:1169–1181. doi: 10.1101/gr.5235706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cootes AP, Muggleton SH, Sternberg MJE. The identification of similarities between biological networks: application to the metabolome and interactome. J. Mol. Biol. 2007;369:1126–1139. doi: 10.1016/j.jmb.2007.03.013. [DOI] [PubMed] [Google Scholar]
- 11.Venkatesan K, Rual JF, Vazquez A, Stelzl U, Lemmens I, Hirozane-Kishikawa T, Hao T, Zenkner M, Xin X, Goh KI, et al. An empirical framework for binary interactome mapping. Nat. Methods. 2009;6:83–90. doi: 10.1038/nmeth.1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pache RA, Aloy P. A novel framework for the comparative analysis of biological networks. PLoS ONE. 2012;7:e31220. doi: 10.1371/journal.pone.0031220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.UniProt-Consortium. The Universal Protein Resource (UniProt) in 2010. Nucleic Acids Res. 2010;38:D142–D148. doi: 10.1093/nar/gkp846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Güldener U, Münsterkötter M, Oesterheld M, Pagel P, Ruepp A, Mewes H-W, Stümpflen V. MPact: the MIPS protein interaction resource on yeast. Nucleic Acids Res. 2006;34:D436–D441. doi: 10.1093/nar/gkj003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ruepp A, Waegele B, Lechner M, Brauner B, Dunger-Kaltenbach I, Fobo G, Frishman G, Montrone C, Mewes H-W. CORUM: the comprehensive resource of mammalian protein complexes–2009. Nucleic Acids Res. 2010;38:D497–D501. doi: 10.1093/nar/gkp914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Singh R, Xu J, Berger B. Global alignment of multiple protein interaction networks with application to functional orthology detection. Proc. Natl Acad. Sci. USA. 2008;105:12763–12768. doi: 10.1073/pnas.0806627105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sharan R, Suthram S, Kelley RM, Kuhn T, McCuine S, Uetz P, Sittler T, Karp RM, Ideker T. Conserved patterns of protein interaction in multiple species. Proc. Natl Acad. Sci. USA. 2005;102:1974–1979. doi: 10.1073/pnas.0409522102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barabasi AL, Oltvai ZN. Network biology: understanding the cell's functional organization. Nat. Rev. Genet. 2004;5:101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- 20.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smoot ME, Ono K, Ruscheinski J, Wang P-L, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2010;27:431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lopes CT, Franz M, Kazi F, Donaldson SL, Morris Q, Bader GD. Cytoscape Web: an interactive web-based network browser. Bioinformatics. 2010;26:2347–2348. doi: 10.1093/bioinformatics/btq430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aranda B, Achuthan P, Alam-Faruque Y, Armean I, Bridge A, Derow C, Feuermann M, Ghanbarian AT, Kerrien S, Khadake J, et al. The IntAct molecular interaction database in 2010. Nucleic Acids Res. 2010;38:D525–D531. doi: 10.1093/nar/gkp878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ceol A, Chatr Aryamontri A, Licata L, Peluso D, Briganti L, Perfetto L, Castagnoli L, Cesareni G. MINT, the molecular interaction database: 2009 update. Nucleic Acids Res. 2010;38:D532–D539. doi: 10.1093/nar/gkp983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Keshava Prasad TS, Goel R, Kandasamy K, Keerthikumar S, Kumar S, Mathivanan S, Telikicherla D, Raju R, Shafreen B, Venugopal A, et al. Human Protein Reference Database–2009 update. Nucleic Acids Res. 2009;37:D767–D772. doi: 10.1093/nar/gkn892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Breitkreutz B-J, Stark C, Reguly T, Boucher L, Breitkreutz A, Livstone M, Oughtred R, Lackner DH, Bähler J, Wood V, et al. The BioGRID Interaction Database: 2008 update. Nucleic Acids Res. 2008;36:D637–D640. doi: 10.1093/nar/gkm1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chautard E, Ballut L, Thierry-Mieg N, Ricard-Blum S. MatrixDB, a database focused on extracellular protein-protein and protein-carbohydrate interactions. Bioinformatics. 2009;25:690–691. doi: 10.1093/bioinformatics/btp025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goll J, Rajagopala SV, Shiau SC, Wu H, Lamb BT, Uetz P. MPIDB: the microbial protein interaction database. Bioinformatics. 2008;24:1743–1744. doi: 10.1093/bioinformatics/btn285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lynn DJ, Winsor GL, Chan C, Richard N, Laird MR, Barsky A, Gardy JL, Roche FM, Chan THW, Shah N, et al. InnateDB: facilitating systems-level analyses of the mammalian innate immune response. Mol. Syst. Biol. 2008;4:218. doi: 10.1038/msb.2008.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Salwinski L, Miller CS, Smith AJ, Pettit FK, Bowie JU, Eisenberg D. The Database of Interacting Proteins: 2004 update. Nucleic Acids Res. 2004;32:D449–D451. doi: 10.1093/nar/gkh086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bader GD, Hogue CWV. Analyzing yeast protein-protein interaction data obtained from different sources. Nat. Biotechnol. 2002;20:991–997. doi: 10.1038/nbt1002-991. [DOI] [PubMed] [Google Scholar]
- 32.Kelley BP, Sharan R, Karp RM, Sittler T, Root DE, Stockwell BR, Ideker T. Conserved pathways within bacteria and yeast as revealed by global protein network alignment. Proc. Natl Acad. Sci. USA. 2003;100:11394–11399. doi: 10.1073/pnas.1534710100. [DOI] [PMC free article] [PubMed] [Google Scholar]