Abstract
Protein lysine acetylation plays an important role in the normal functioning of cells, including gene expression regulation, protein stability and metabolism regulation. Although large amounts of lysine acetylation sites have been identified via large-scale mass spectrometry or traditional experimental methods, the lysine (K)-acetyl-transferase (KAT) responsible for the acetylation of a given protein or lysine site remains largely unknown due to the experimental limitations of KAT substrate identification. Hence, the in silico prediction of KAT-specific acetylation sites may provide direction for further experiments. In our previous study, we developed the acetylation set enrichment based (ASEB) computer program to predict which KAT-families are responsible for the acetylation of a given protein or lysine site. In this article, we provide KAT-specific acetylation site prediction as a web service. This web server not only provides the online tool and R package for the method in our previous study, but several useful services are also included, such as the integration of protein–protein interaction information to enhance prediction accuracy. This web server can be freely accessed at http://cmbi.bjmu.edu.cn/huac.
INTRODUCTION
Protein lysine acetylation is one of the most important post-translational modifications; it plays important roles in protein stability, gene expression regulation, protein–protein interactions (PPI) and cellular metabolism (1–4). Acetylation was first discovered in histones (5) and subsequently, in non-histone proteins (6). The number of acetylated proteins is rapidly increasing because of the development of high-throughput technologies, such as immune-precipitation combined mass spectrometric analysis (7). Over 2000 human acetylated proteins (4000 lysine sites) have been identified via traditional experimental methods and large-scale mass spectrometric analysis (8). However, determining which lysine (K)-acetyl-transferases (KATs) are responsible for the acetylation of given proteins remains difficult.
Similar to kinases that catalyze the phosphorylation of a specific subset of substrates, KATs are substrate-specific enzymes. Over 20 human KATs have been identified (9). Although categorizing all of these KATs remains difficult due to their variety, nine KATs have been divided into three families based on sequence and structural similarity (10). The families include CBP/p300, GCN5/PCAF and MYST. Each of the three families catalyzes a special subgroup of substrates. For instance, WRN is acetylated by CBP/p300, but not by PCAF or TIP60 (11). Although different KAT families can target the same protein, they possibly acetylate different lysine sites. For instance, PCAF acetylates p53 at site K320 (12), whereas TIP60 acetylates the same protein at K120 (13). Thus, the construction of in silico tools for the prediction of KAT-specific acetylation sites is reasonable.
Previous in silico prediction methods have focused on acetylation itself. For example, several research groups have predicted acetylation using the support vector machine (SVM) method. Such methods can classify new sequences by learning the features of sequence contexts surrounding the acetylated lysines (14–16). Aside from SVM, other methods, such as meta-analysis and sequence clustering, have also been used to predict acetylation sites (17,18). However, these tools can only provide information on the probability of acetylation for a given protein, but cannot provide KAT-specific acetylation prediction.
In this study, we constructed a KAT-specific prediction server called acetylation set enrichment based (ASEB), which can predict not only the acetylation state, but also the responsible KAT family. The prediction method was initially established based on the sequence similarity principle. Detailed algorithms were presented in our previous study (8). In this article, we further integrated protein–protein interaction information to enhance prediction accuracy. A graph of the rank of the predicted P-value from the background P-values was also provided, which would enable users to evaluate the predicted results. We also evaluated the sensitivity and specificity of ASEB on a new independent data set, which proves that the web server could work for other species besides humans. We take the human protein TP53 as an example to illustrate the utility of the ASEB server.
WEB SERVER CONSTRUCT
The ASEB web server takes advantage of Asynchronous JavaScript and XML technology, which can dynamically display the status of the predicting process. The web site follows HTML 4.01 Strict and CSS version 2.1 standards to maintain consistency across different browsers. Compared with the method in the original study (8), the web server was improved in three areas as follows. (i) The enzyme often interacts with its substrates directly or indirectly through scaffold proteins to achieve a reaction. PPI information is helpful in predicting enzyme-mediated biological reactions; as such, the shortest path between KAT and query protein in the protein–protein interaction network was added to enhance the accuracy of prediction. (ii) Suggestions on cut-off selection may be useful for users of the web server; thus, we graphed the rank of the predicted P-value from the background P-values, which would enable users to evaluate predicted results. (iii) Although the web server was constructed based on human proteins, it can also be used to predict acetylated proteins from other species due to the evolutional conservation of acetylated proteins and KATs. Acetylation data from other species were further added and the performance on this data was evaluated, revealing that the web server could also work on these species.
Shortest path between KAT and query protein
In the acetylation process, the enzyme interacts with its substrates directly or indirectly. We could hypothesize that if KATs acetylate the query protein; they should interact directly or at least be mediated by the scaffold proteins. Research has shown that PPI information can enhance the accuracy of protein kinase substrate prediction (19). In this article, we provided the shortest path between KAT and the query protein in the PPI network. The prediction results should be more reliable when the KAT and the query protein interact. The shortest path between the query protein and a KAT was calculated via Dijkstra's algorithm. During the determination of the shortest path, the values of the edges in the PPI network were estimated via the following two methods: (i) the values of the edges were assigned as one for the PPI edges obtained from the database PINA (20) and (ii) the value of the edge between ‘protein1’ and ‘protein2’ was assigned as ‘log(1000 − combinedScore(protein1, protein2))’ for the PPI edges obtained from the database STRING (21). The combined scores between proteins were queried from the database STRING. The service of shortest path in the PPI network between the KAT and the query protein is currently limited to ‘Homo sapiens’ proteins.
Cut-off selection
With ASEB, each query lysine site can get a P-value. A lower P-value indicates a higher probability of the acetylation of the lysine site by the selected KAT family. Furthermore, each P-value's rank in the background P-values was generated. The background P-values were calculated from all lysine sites on human proteins and then ranked from lowest to highest. Once a P-value is given, its rank in the background can be determined, which would be useful for users to evaluate predicted results. To control the false-positive predictions, we suggest users pay more attention to the lysine sites with P-values lower than the top 10%; in this case, the estimated specificity will be higher than 90%. Lysine sites passing the suggested cut-off are highlighted by color in the table of prediction results on the web site. The background set (all lysine sites on human proteins) should contain unreported acetylation sites; as such, the specificity is very likely underestimated. In our opinion, this cut-off should be loosened once interaction between KAT and query protein occurs. In applications, users can adjust the cut-off values according to the trade-off between discovering more putative acetylation sites and making fewer false-positive predictions.
Evaluation on new independent data set
In our previous study, we initially ran a leave-one-out method to estimate the sensitivity for known acetylated sites and perform the ASEB method on 1000 randomly selected lysine sites to estimate specificity. The detailed results are available on the web server. We also predicted the acetylation states of proteins without known KATs and experimentally validated the prediction results. In this study, we further tested the method on a new independent data set from other species. This data set contains 42 and 30 known acetylation sites from other species, which are acetylated by the orthologs of CBP/p300 and GCN5/PCAF. All these data can be accessed from the web server. A total of 1000 lysine sites were randomly selected from other species as a background data set. The test results indicated that the ASEB method performed similarly well on other species; the detailed results can be found in Table 1. Given that this background test set might still contain unreported acetylation sites, the specificity estimated with this test set might be underestimated.
Table 1.
Validation results
| KAT Family | CBP/p300 |
GCN5/PCAF |
||
|---|---|---|---|---|
| Known (Sensitivity) | Background (Specificity) | Known (Sensitivity) | Background (Specificity) | |
| Total peptides | 42 | 1000 | 30 | 1000 |
| P ≤ 1e−4 | 4 (9.5%) | 18 (98.2%) | 8 (26.7%) | 4 (99.6%) |
| P ≤ 1e−3 | 8 (19.0%) | 32 (96.8%) | 13 (43.3%) | 9 (99.1%) |
| P ≤ 1e−2 | 11 (26.2%) | 77 (92.3%) | 16 (53.3%) | 35 (96.5%) |
| P ≤ 1e−1 | 25 (59.5%) | 181 (81.9%) | 20 (66.7%) | 137 (86.3%) |
Input
To predict whether lysine sites within a given protein sequence can be acetylated by specific KAT families, users should initially select the KAT family that interests them. The query protein sequence can then be directly input into the ‘Sequences’ text box. Other input terms, such as Swiss-Prot accession number or protein name, are also permitted but are limited to ‘Homo sapiens’. If the accession number or protein name is available, users can click the ‘Load Sequence’ button to obtain the protein sequence instead of using manual input. When users predict acetylation sites on protein sequences from other species, the predicted KATs should be correspondingly translated into KATs in their own species. For example, a sequence from yeast predicted to be acetylated by the GCN5/PCAF family should be translated as acetylated by the yGcn5 protein, instead of by the human GCN5 or PCAF protein. Aside from the prediction of the acetylation state based on the sequence, the interaction network between KATs and the query protein can also be generated by entering the protein accession number or name.
On the prediction page, the web server takes the TP53 with a Swiss-Prot accession number P04637 as an input example. An ‘Example’ button is provided to load this protein. If users click this button, the accession number for TP53 will be entered automatically. If users subsequently click the ‘Load Sequence’ button, the protein sequence would appear in the ‘Sequences’ text box.
Output
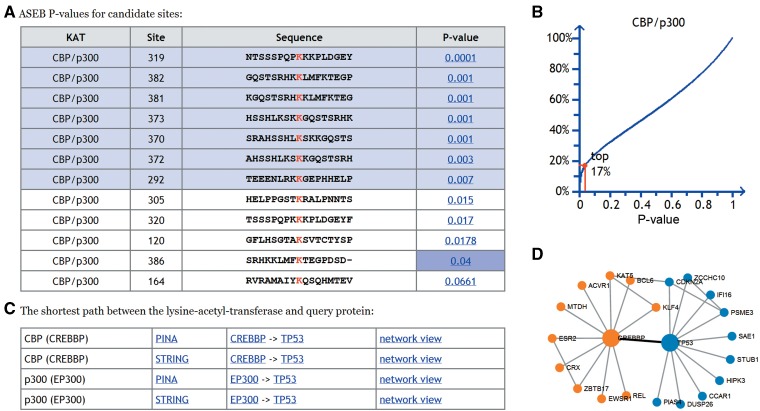
The outputs include the prediction results for each lysine site and the PPI interaction networks between the query protein and the KATs (Figure 1). The prediction results are shown in a table containing four columns: the selected KAT family, the lysine position, the 17 length peptides surrounding the lysine and the ASEB P-values (Figure 1A). The range of the P-value is from 0.0001 to 1, with an interval of 0.0001. For details on the algorithm, refer (8). If users click on a P-value, a plot will appear and show the rank of the P-value from the background P-values for all lysine sites on human proteins (Figure 1B). Lysine sites with P-values ranked in the top 10% are highlighted by color in the table of prediction results (Figure 1A).
Figure 1.
Prediction results for human protein TP53. (A) Table containing predicted P-values for each lysine site on TP53. The sites with lower P-values in the top 10% were highlighted by background color. (B) Plot showing the rank of P-value 0.04 from the background. The X-axis indicates the P-value, and the Y-axis indicates the rank of this P-value. (C) Shortest path between the lysine-acetyl-transferase (CREBBP) and the query protein (TP53). (D) Example of the PPI network view. The shortest path between the lysine-acetyl-transferase (CREBBP) and the query protein (TP53) is highlighted by the bold black line.
When users provide a Swiss-Prot accession number or protein name, the PPI interaction networks will be presented. Various paths exist from the KATs to the input proteins, and the shortest of these was extracted to represent the relationship between the interacting proteins. These networks were derived from two main PPI databases, PINA and STRING (20,21). For example, when we selected the CBP/p300 family and submitted the accession number for TP53, we obtained the shortest path between the CBP or p300 and the TP53 in the PPI network (Figure 1C). Both CBP and p300 directly interact with TP53. By clicking the network view link, users can visualize the interaction between CBP or p300 and TP53 (Figure 1D). The shortest path from the KAT to the query protein is highlighted by a bold black line. A step-by-step tutorial page with detailed explanation can be accessed on the web server.
Other services provided by the ASEB web server
In addition to KAT-specific acetylation prediction, users can also search for the identified acetylation proteins and sites that have been collected. Search results include acetylated lysine sites on query proteins, KATs responsible for the lysine sites (if present), and the PubMed ID for original papers describing the corresponding acetylation. These data can also be downloaded directly. A template program in Perl was also provided to allow users to access the services programmatically, rather than through manual interaction. Users who are familiar with R can download the ASEB R package (developing version) from Bioconductor. The R package can process a large number of proteins simultaneously. The instructions for use are included in the package.
CONCLUSION
ASEB is a web server for KAT-specific acetylation prediction that is available for free. The efficacy of ASEB is validated via independent methods, including biological experiments. To our knowledge, ASEB is the first KAT-specific acetylation prediction method. Although the present version only predicts acetylation catalyzed by two KAT families, it is helpful for scientists in the acetylation field. We will continue to collect experimentally identified acetylation sites and responsible KATs. We will update the web server when the number of newly detected acetylation sites is adequate to enhance efficiency, and when data from other KAT families can provide reliable predicted results.
FUNDING
The National Basic Research Program [2011CBA01104]; the National High-tech R&D Program [2012AA020401] of China, as well as the National Natural Science Foundation of China [60905014 and 31030041]. The funding agencies played no active role in the study design, data collection, analysis, decision to publish or preparation of the manuscript. Funding for open access charge: the National Natural Science Foundation of China [60905014].
Conflict of interest statement. None declared.
REFERENCES
- 1.Das C, Kundu TK. Transcriptional regulation by the acetylation of nonhistone proteins in humans: a new target for therapeutics. IUBMB Life. 2005;57:137–149. doi: 10.1080/15216540500090629. [DOI] [PubMed] [Google Scholar]
- 2.Kim SC, Sprung R, Chen Y, Xu Y, Ball H, Pei J, Cheng T, Kho Y, Xiao H, Xiao L, et al. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol. Cell. 2006;23:607–618. doi: 10.1016/j.molcel.2006.06.026. [DOI] [PubMed] [Google Scholar]
- 3.Spange S, Wagner T, Heinzel T, Kramer OH. Acetylation of non-histone proteins modulates cellular signalling at multiple levels. Int. J. Biochem. Cell Biol. 2009;41:185–198. doi: 10.1016/j.biocel.2008.08.027. [DOI] [PubMed] [Google Scholar]
- 4.Zhao S, Xu W, Jiang W, Yu W, Lin Y, Zhang T, Yao J, Zhou L, Zeng Y, Li H, et al. Regulation of cellular metabolism by protein lysine acetylation. Science. 2010;327:1000–1004. doi: 10.1126/science.1179689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Allfrey VG, Faulkner R, Mirsky AE. Acetylation and methylation of histones and their possible role in the regulation of RNA synthesis. Proc. Natl Acad. Sci. USA. 1964;51:786–794. doi: 10.1073/pnas.51.5.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gu W, Roeder RG. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 7.Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 8.Li T, Du Y, Wang L, Huang L, Li W, Lu M, Zhang X, Zhu WG. Characterization and prediction of lysine (K)-acetyl-transferase specific acetylation sites. Mol. Cell Proteom. 2012;11:M111. 011080. doi: 10.1074/mcp.M111.011080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roth SY, Denu JM, Allis CD. Histone acetyltransferases. Annu. Rev. Biochem. 2001;70:81–120. doi: 10.1146/annurev.biochem.70.1.81. [DOI] [PubMed] [Google Scholar]
- 10.Marmorstein R, Roth SY. Histone acetyltransferases: function, structure, and catalysis. Curr. Opin. Genet. Dev. 2001;11:155–161. doi: 10.1016/s0959-437x(00)00173-8. [DOI] [PubMed] [Google Scholar]
- 11.Li K, Wang R, Lozada E, Fan W, Orren DK, Luo J. Acetylation of WRN protein regulates its stability by inhibiting ubiquitination. PLoS One. 2010;5:e10341. doi: 10.1371/journal.pone.0010341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu L, Scolnick DM, Trievel RC, Zhang HB, Marmorstein R, Halazonetis TD, Berger SL. p53 sites acetylated in vitro by PCAF and p300 are acetylated in vivo in response to DNA damage. Mol. Cell. Biol. 1999;19:1202–1209. doi: 10.1128/mcb.19.2.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sykes SM, Mellert HS, Holbert MA, Li K, Marmorstein R, Lane WS, McMahon SB. Acetylation of the p53 DNA-binding domain regulates apoptosis induction. Mol. Cell. 2006;24:841–851. doi: 10.1016/j.molcel.2006.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gnad F, Ren S, Choudhary C, Cox J, Mann M. Predicting post-translational lysine acetylation using support vector machines. Bioinformatics. 2010;26:1666–1668. doi: 10.1093/bioinformatics/btq260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li S, Li H, Li M, Shyr Y, Xie L, Li Y. Improved prediction of lysine acetylation by support vector machines. Protein Pept. Lett. 2009;16:977–983. doi: 10.2174/092986609788923338. [DOI] [PubMed] [Google Scholar]
- 16.Xu Y, Wang XB, Ding J, Wu LY, Deng NY. Lysine acetylation sites prediction using an ensemble of support vector machine classifiers. J. Theor. Biol. 2010;264:130–135. doi: 10.1016/j.jtbi.2010.01.013. [DOI] [PubMed] [Google Scholar]
- 17.Basu A, Rose KL, Zhang J, Beavis RC, Ueberheide B, Garcia BA, Chait B, Zhao Y, Hunt DF, Segal E, et al. Proteome-wide prediction of acetylation substrates. Proc. Natl Acad. Sci. USA. 2009;106:13785–13790. doi: 10.1073/pnas.0906801106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schwartz D, Chou MF, Church GM. Predicting protein post-translational modifications using meta-analysis of proteome scale data sets. Mol. Cell Proteom. 2009;8:365–379. doi: 10.1074/mcp.M800332-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Linding R, Jensen LJ, Ostheimer GJ, van Vugt MA, Jorgensen C, Miron IM, Diella F, Colwill K, Taylor L, Elder K, et al. Systematic discovery of in vivo phosphorylation networks. Cell. 2007;129:1415–1426. doi: 10.1016/j.cell.2007.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cowley MJ, Pinese M, Kassahn KS, Waddell N, Pearson JV, Grimmond SM, Biankin AV, Hautaniemi S, Wu J. PINA v2.0: mining interactome modules. Nucleic Acids Res. 2012;40:D862–D865. doi: 10.1093/nar/gkr967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Szklarczyk D, Franceschini A, Kuhn M, Simonovic M, Roth A, Minguez P, Doerks T, Stark M, Muller J, Bork P, et al. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011;39:D561–D568. doi: 10.1093/nar/gkq973. [DOI] [PMC free article] [PubMed] [Google Scholar]