Abstract
The Protein Identifier Cross-Reference (PICR) service is a tool that allows users to map protein identifiers, protein sequences and gene identifiers across over 100 different source databases. PICR takes input through an interactive website as well as Representational State Transfer (REST) and Simple Object Access Protocol (SOAP) services. It returns the results as HTML pages, XLS and CSV files. It has been in production since 2007 and has been recently enhanced to add new functionality and increase the number of databases it covers. Protein subsequences can be Basic Local Alignment Search Tool (BLAST) against the UniProt Knowledgebase (UniProtKB) to provide an entry point to the standard PICR mapping algorithm. In addition, gene identifiers from UniProtKB and Ensembl can now be submitted as input or mapped to as output from PICR. We have also implemented a ‘best-guess’ mapping algorithm for UniProt. In this article, we describe the usefulness of PICR, how these changes have been implemented, and the corresponding additions to the web services. Finally, we explain that the number of source databases covered by PICR has increased from the initial 73 to the current 102. New resources include several new species-specific Ensembl databases as well as the Ensembl Genome ones. PICR can be accessed at http://www.ebi.ac.uk/Tools/picr/.
INTRODUCTION
Mapping protein identifiers across different data sources is a difficult task. Each major protein database [UniProt (1), Ensembl (2), RefSeq (3), etc] assigns identifiers to protein sequences according to their own internal guidelines and identifier pattern. While the major data providers try and maintain cross-references to each other, different release schedules make it hard to keep the data synchronized and this is made even more difficult when trying to keep track of novel or proprietary databases. The complexity of the problem is compounded by database redundancy, where identical protein sequences can be assigned different accessions, and by the fact that identifiers and sequences can change due to new data or predictive algorithms. Protein sequence databases are always in flux, and this has a significant impact on the quality and long-term reliability of submission-driven data repositories (4).
There have been attempts to create unified identifier schemes in the past. Both Life Science Identifiers (LSID) (5) and Sequence Globally Unique Identifiers (SEGUID) (6) attempted to solve the identifiers problem by assigning an identifier that is unique to a given protein sequence. However, neither of these has achieved widespread adoption.
There are a few tools that attempted to solve the protein identifier mapping problem. Unfortunately, most of these are limited either by the scope of the protein database they cover, by being limited to a single species, or by having limited usability (requiring periodic maintenance to update the local databases, lack of support for programmatic requests, etc). For instance, IdConverter provides mapping between a significant number of databases from the gene level to the functional level but it is restricted to three species: human, mouse and rat. In addition, all of the databases that it uses are quite out of date (7). CaBIG GeneConnect (https://cabig.nci.nih.gov/tools/GeneConnect/), from the National Cancer Institute, provides mappings both programmatically and interactively, but has not been updated since 2007, and maps between a limited number of databases. Protein Information Resource's (PIR) ID-mapping service allows direct identifier mapping between a small numbers of databases (http://pir.georgetown.edu/pirwww/search/idmapping.shtml). MatchMiner, also by the National Cancer Institute, has good support for gene names but is limited to human and mouse genes (8). Wayne State's Onto-Translate (http://vortex.cs.wayne.edu/projects.htm#Onto-Translate) is gene focused and requires a login to use. ‘The Synergizer’ supports both gene and protein ID mapping, but has a limited set of databases and no programmatic access is provided (http://llama.mshri.on.ca/synergizer/translate/). The Gene ID conversion tool of the Database for Annotation, Visualization and Integrated Discovery (DAVID) bioinformatics resources (9), makes use of the concept of DAVID ‘gene’, to support identifier mappings between around 25 different gene and protein resources, but offers only limited programmatic access. Finally, the UniProt ID-mapping system (http://www.uniprot.org/mapping/) supports identifier mapping among a high number of resources. Its main limitations are that both the specified source and target identifier database are unique per query, and that protein sequences are not supported.
The Protein Identifier Cross-Reference (PICR) service was originally developed in 2007 at the European Bioinformatics Institute (EBI) to address the shortcomings described above (10). PICR is a web application, accessible at http://www.ebi.ac.uk/Tools/picr/, which maps active and deprecated protein and gene identifiers, complete amino acid sequences and protein subsequences to the corresponding identifiers in over 100 different databases. PICR can be accessed interactively through a website and programmatically through Representational State Transfer (REST) and Simple Object Access Protocol (SOAP) interfaces. It can map accessions from several sources to multiple target databases in one request. Mappings can be limited to specific taxa or across all species. Since PICR's release in 2007, it has been very heavily used, averaging more than 12.5 million monthly hits since going into production, with upwards of 90% of requests coming through the web services. PICR is used routinely by other EBI resources such as Proteomics Identifications database (PRIDE) (11) and IntAct (12), as an essential part of their data workflow, and it has also demonstrated its usefulness as a data analysis tool (4,13).
In this article, we describe recent additions to PICR allowing the identification of protein subsequences using the Basic Local Alignment Search Tool (BLAST) as well as the addition of mappings from gene identifiers to any of the databases referenced in UniProt Archive (UniParc), and the addition of a UniProt ‘best guess’ option. An online user guide is available at http://www.ebi.ac.uk/Tools/picr/userguide.do.
IMPLEMENTATION: NEW FEATURES
BLAST support
The implementation of PICR has been described in depth before (10). In summary, PICR originally allowed submission of complete protein sequences and protein identifiers via one of three interfaces: programmatically using SOAP or REST services, or interactively using the website. Mapping by sequence is done by computing a CRC64 checksum of the sequence and matching it to a checksum in the UniParc (14) database to obtain a unique UniParc protein identifier (UPI). This UPI is then used to query the UniParc cross-reference table (xref) for identifiers matching the desired search criteria (target database, active versus deleted status, taxonomy, etc). Mapping by accession is done in a similar manner, with the xref table as an entry point to retrieve a list of all UPIs that have been associated with a given protein identifier now and in the past (10).
Since sequence-based mapping was previously done by taking a checksum of the submitted sequence, any variation from the canonical sequence would change the checksum and produce different, and often limited, results. This represented a significant problem for any users with any data set that contains either a subsection of the complete sequence, or any slight change from already known sequences. To alleviate this problem, support for BLAST searching was added to the web site front end, as well as to the REST and SOAP web services. To achieve this goal, we used the existing BLAST web service provided by the EBI (15) (http://www.ebi.ac.uk/Tools/webservices/services/sss/ncbi_blast_soap). In order to optimize the process and minimize query times, BLAST queries were parallelized. The EBI service allows up to 25 queries at once, which substantially reduced the search time.
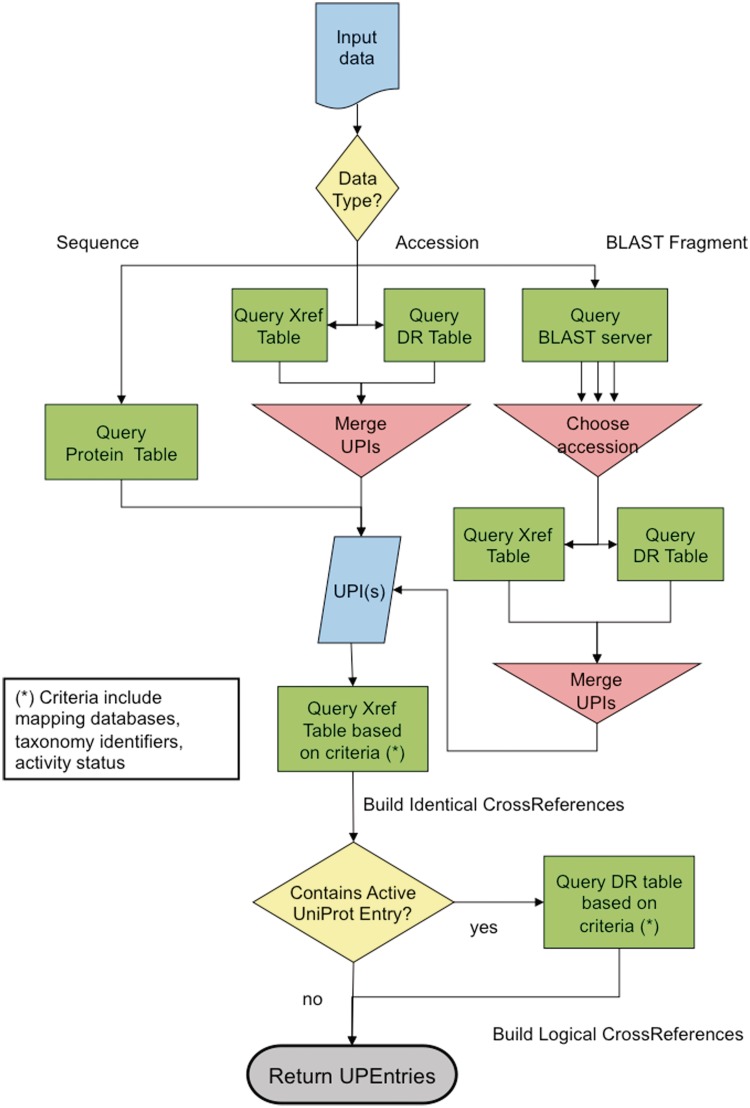
The PICR BLAST algorithm takes a list of protein sequences and the parameters to be passed to the EBI BLAST service, such as the number of hits to return, species and identity value filters for the return values, as well as other various BLAST algorithm parameters options. The BLAST search is done in parallel for each protein sequence and the results are collated once the individual queries are finished. The user is then given the option of selecting which one of the BLAST results for each protein sequence can be used as an entry point to the mapping-by-accession algorithm. If one of the web service interfaces is used, the first, best ranked BLAST hit is automatically used. A flowchart of the updated mapping algorithm is shown in Figure 1.
Figure 1.
The updated PICR mapping algorithm. The first part of the algorithm deals with finding all UniParc entries that pertain to the source data, be it coming from protein sequences or identifiers. From there, the UniParc entries are used to build cross-references to the required target databases, based on the initial search criteria.
The addition of BLAST functionality required both the interactive and programmatic interfaces of PICR to be updated to capture the BLAST-specific options and gracefully handle the new workflow generated by the necessity to present the BLAST results to the user (Figure 2). JavaScript functions were written to check all of the BLAST input as well as to propagate ‘Limit by species’ to the BLAST search form. BLAST support was added to the SOAP and REST interfaces by updating the web service descriptor (http://www.ebi.ac.uk/Tools/picr/service?wsdl) to handle new messages as required by the service (getUPIForBlastSequence and getUPIForBlastSequenceResponse) and to enhance the object model to be able to capture all the required BLAST parameters. It should be noted that programmatic calls include only one input sequence and must include which BLAST database to search against, the minimum identity value to accept as a response, the species taxon to filter on, in which UniParc databases to look for cross-references, how to filter results and a set of BLAST parameters to use. The RESTful interface to PICR uses the same XML service descriptor as the SOAP service to generate output messages so that interface only needed to be able to handle the new input parameters to extend the functionality it provides. Full documentation and some template code are available at http://www.ebi.ac.uk/Tools/picr/WSDLDocumentation.do (for the SOAP web service) and http://www.ebi.ac.uk/Tools/picr/RESTDocumentation.do (for the REST web service).
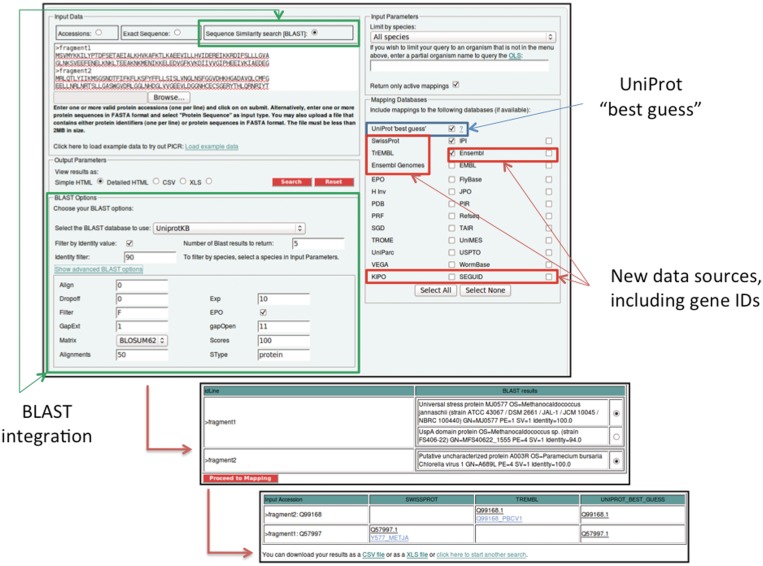
Figure 2.
The updated PICR web interface. The integration of BLAST support to PICR required an additional step to the identifier mapping process. Users can select which BLAST hit to use as an entry point to the map-by-accession PICR mapping algorithm. The final result that is presented to the user will contain both the definition line for each sequence and the selected UniProt accession number, so that the user can keep track of what was originally submitted for mapping.
Improved database coverage and further support for gene identifiers
More than 30 new source protein databases have been made available for mapping since PICR went into production in 2007. As of January 2012, the current number of supported resources is 102. Among the additions are, the Korean Intellectual Property Office (KIPO) database, the five existing Ensembl Genome databases (bacteria, fungi, metazoa, plants and protists, which cover over 335 organisms, as of January 2012) (16) and upwards of 20 Ensembl species-specific databases (listed in the Supplementary Table S1). Functionality was also implemented to compute SEGUIDs based on the UniParc protein sequences (though it is not currently possible to use SEQUIDs as search input parameters).
Gene-to-protein identifier mapping for Ensembl, Ensembl Genomes and UniProt gene identifiers has also been implemented in the latest version of the service. This allows users to map to and from gene identifiers. The gene identifiers are parsed from the latest UniProt release files [GN lines for UniProt gene identifiers and DR lines for the Ensembl and Ensembl Genomes gene identifiers (17)] and will be returned as logical cross-references in the PICR data model.
UniProt ‘best guess’ mapping algorithm
It is often the case that, when trying to map to the UniProt KnowledgeBase (UniProtKB) identifiers, a search term will generate several UniProtKB/Swiss-Prot (manually curated subset of UniProtKB) and UniProtKB/TrEMBL (UniProtKB subset based on the automatic annotation) matches. It is then tricky for the users to select the best option, in particular, in an automated workflow. We produced a novel search algorithm that, given the output of a standard PICR search, would generate a single UniProt accession number corresponding to the best match for the submitted accession. The UniProt ‘best guess’ option is defined as the entry with the longest protein sequence in the following subsets within UniProtKB, by order of preference, Swiss-Prot canonical sequence, Swiss-Prot annotated isoform, TrEMBL canonical sequence and TrEMBL annotated isoform.
DISCUSSION
Since being put in production, in 2007, PICR has gone a long way to make a difficult, labour-intensive problem easier to manage and automate by providing multiple interfaces to map protein identifiers, sequences and more recently gene identifiers. By fulfilling all of its original use cases, of offering both programmatic and interactive interfaces, allowing mappings to and from several databases at once and allowing batch queries, PICR has become the most used service provided by the EBI Proteomics Services Team. New source databases are constantly being integrated into the UniProt Archive (UniParc), and by the same token into PICR, always increasing the scope and breadth of coverage. Responding to user-generated feedback, we have introduced new functionality to PICR that will make it even more versatile. The UniProt ‘best guess’ option will remove complexity in cases where a simple, single mapping to UniProt is required. Mapping by sequence similarity using BLAST will greatly improve the relevance of PICR hits for sequence fragments or where the possibility of sequence errors, artefacts or regions of high variability exists. We will continue to increase database coverage as more resources become available and should users wish to discuss requests for new functionality; the authors wholeheartedly encourage them to contact the PICR helpdesk with their suggestions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table 1.
FUNDING
EU FP7 grant SLING [226073 to R.C.]; EU FP7 grants LipidomicNet [202272 to J.A.V.]; ProteomeXchange [260558 to J.A.V.]; EU FP7 PSIMEx grant [FP7-HEALTH-2007-223411 to M.D.]; Wellcome Trust [WT085949MA to F.R.]. Funding for open access charge: EU FP7 ProteomeXchange grant [260558].
Conflict of interest statement. None declared.
REFERENCES
- 1.The UniProt Consortium. Ongoing and future developments at the Universal Protein Resource. Nucleic Acids Res. 2011;39:D214–D219. doi: 10.1093/nar/gkq1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Flicek P, Amode MR, Barrell D, Beal K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fairley S, Fitzgerald S, et al. Ensembl 2012. Nucleic Acids Res. 2012;40:D84–D90. doi: 10.1093/nar/gkr991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pruitt KD, Tatusova T, Brown GR, Maglott DR. NCBI Reference Sequences (RefSeq): current status, new features and genome annotation policy. Nucleic Acids Res. 2012;40:D130–D135. doi: 10.1093/nar/gkr1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Griss J, Cote RG, Gerner C, Hermjakob H, Vizcaino JA. Published and perished? The influence of the searched protein database on the long-term storage of proteomics data. Mol. Cell Proteomics. 2011;10:M111 008490. doi: 10.1074/mcp.M111.008490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clark T, Martin S, Liefeld T. Globally distributed object identification for biological knowledgebases. Brief Bioinform. 2004;5:59–70. doi: 10.1093/bib/5.1.59. [DOI] [PubMed] [Google Scholar]
- 6.Babnigg G, Giometti CS. A database of unique protein sequence identifiers for proteome studies. Proteomics. 2006;6:4514–4522. doi: 10.1002/pmic.200600032. [DOI] [PubMed] [Google Scholar]
- 7.Alibes A, Yankilevich P, Canada A, Diaz-Uriarte R. IDconverter and IDClight: conversion and annotation of gene and protein IDs. BMC Bioinformatics. 2007;8:9. doi: 10.1186/1471-2105-8-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bussey KJ, Kane D, Sunshine M, Narasimhan S, Nishizuka S, Reinhold WC, Zeeberg B, Ajay W, Weinstein JN. MatchMiner: a tool for batch navigation among gene and gene product identifiers. Genome Biol. 2003;4:R27. doi: 10.1186/gb-2003-4-4-r27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 10.Cote RG, Jones P, Martens L, Kerrien S, Reisinger F, Lin Q, Leinonen R, Apweiler R, Hermjakob H. The Protein Identifier Cross-Referencing (PICR) service: reconciling protein identifiers across multiple source databases. BMC Bioinformatics. 2007;8:401. doi: 10.1186/1471-2105-8-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vizcaino JA, Cote R, Reisinger F, Barsnes H, Foster JM, Rameseder J, Hermjakob H, Martens L. The Proteomics Identifications database: 2010 update. Nucleic Acids Res. 2010;38:D736–D742. doi: 10.1093/nar/gkp964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kerrien S, Aranda B, Breuza L, Bridge A, Broackes-Carter F, Chen C, Duesbury M, Dumousseau M, Feuermann M, Hinz U, et al. The IntAct molecular interaction database in 2012. Nucleic Acids Res. 2012;40:D841–D846. doi: 10.1093/nar/gkr1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Griss J, Martin M, O'Donovan C, Apweiler R, Hermjakob H, Vizcaino JA. Consequences of the discontinuation of the International Protein Index (IPI) database and its substitution by the UniProtKB “complete proteome” sets. Proteomics. 2011;11:4434–4438. doi: 10.1002/pmic.201100363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leinonen R, Diez FG, Binns D, Fleischmann W, Lopez R, Apweiler R. UniProt archive. Bioinformatics. 2004;20:3236–3237. doi: 10.1093/bioinformatics/bth191. [DOI] [PubMed] [Google Scholar]
- 15.Goujon M, McWilliam H, Li W, Valentin F, Squizzato S, Paern J, Lopez R. A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res. 2010;38:W695–W699. doi: 10.1093/nar/gkq313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kersey PJ, Staines DM, Lawson D, Kulesha E, Derwent P, Humphrey JC, Hughes DS, Keenan S, Kerhornou A, Koscielny G, et al. Ensembl Genomes: an integrative resource for genome-scale data from non-vertebrate species. Nucleic Acids Res. 2012;40:D91–D97. doi: 10.1093/nar/gkr895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.The UniProt Consortium. UniProt Knowledgebase User Manual. 2012 http://www.uniprot.org/manual/ [Google Scholar]