Abstract
The 2012 Bioinformatics Links Directory update marks the 10th special Web Server issue from Nucleic Acids Research. Beginning with content from their 2003 publication, the Bioinformatics Links Directory in collaboration with Nucleic Acids Research has compiled and published a comprehensive list of freely accessible, online tools, databases and resource materials for the bioinformatics and life science research communities. The past decade has exhibited significant growth and change in the types of tools, databases and resources being put forth, reflecting both technology changes and the nature of research over that time. With the addition of 90 web server tools and 12 updates from the July 2012 Web Server issue of Nucleic Acids Research, the Bioinformatics Links Directory at http://bioinformatics.ca/links_directory/ now contains an impressive 134 resources, 455 databases and 1205 web server tools, mirroring the continued activity and efforts of our field.
COMMENTARY
‘Because the number of artifacts has increased greatly, it is impossible for many bioinformatics researchers to track tools, databases, and methods in the field—or even perhaps within their own specialty area. More critically, however, biologist users and scientists approaching the field do not have a comprehensive index of bioinformatics algorithms, databases, and literature annotated with information about their context and appropriate use.’ (1)
The need to not only share bioinformatics databases, tools and resources but also compile a bioinformatics ‘resourceome’ has been evident for some time (1). In dedicating a special issue each year since 1993 to new databases or updates and a second issue each year since 2003 to new web server tools or updates, Nucleic Acids Research has successfully created a viable and recognizable dissemination mechanism. Each year since 2005 and extending back to 2003, the Bioinformatics Links Directory has coordinated with Nucleic Acids Research (NAR) to update their community driven ‘resourceome’ with all of NAR’s web server tools (2–8). This coordinated effort has resulted in a comprehensive public repository of bioinformatics tools and databases, annotated with contextual information and organized by functional relevance (http://bioinformatics.ca/links_directory). Users requiring a bioinformatics tool or database for their research may search the Bioinformatics Links Directory using any number of its features, from browsing the biological categories and their drop-down subcategories on the main page, to computing a keyword search from the search box or selecting related links. Content within the Directory is organized by biological subject, with subcategories of common tasks relevant to each subject listed under the corresponding category (Figure 1). A keyword search from the search box (Figure 2) returns links containing the entered keywords found within either the link’s title, description or tag terms—a search for ‘gene set enrichment’ for example returns links containing a Boolean combination of gene AND set AND enrichment, with these keywords highlighted in yellow where they appear in the returned list (Figure 3). Other specific searches may also be computed such as exact word matches using quotation marks. Returned links from a given search may be refined to one or more of the link types within the Links Directory (tools, resources or databases), using the ‘Search Options’ feature (Figure 4). From a link of interest, related or similar links within the Links Directory are retrievable by selecting the ‘Related Links’ tab, where related links are chosen based on the PubMed-related citations feature for the primary link. To aid with determining the usefulness and currency of a link, all links within the Links Directory with one or more citations have a per PubMed Identifier (PMID) and cumulative Google Scholar (9) citation count, as well as a Google+ and Twitter sharing count (10) (Figure 5). A keyword search will return links in descending order of cumulative citation count. Detailed explanations and examples of searching within the Links Directory are provided on the tutorials page at http://www.bioinformatics.ca/links_directory/tutorial. These annotations and search features provide the ‘biologist user with information about a link’s context’ and usefulness so to narrow down the algorithm or database within the Bioinformatics Links Directory ‘resourceome’ best suited to their research need (1).
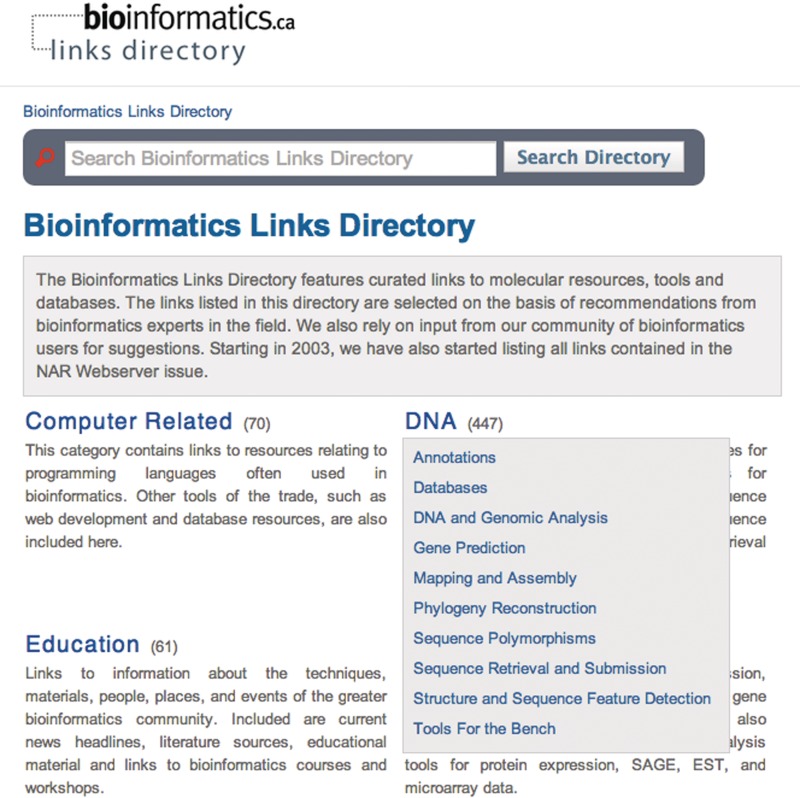
Figure 1.
The homepage of the Bioinformatics Links Directory displays the functional categories and subcategories within which more than 1700 tools, resources and databases are contained. The DNA category is shown expanded into its subcategories.
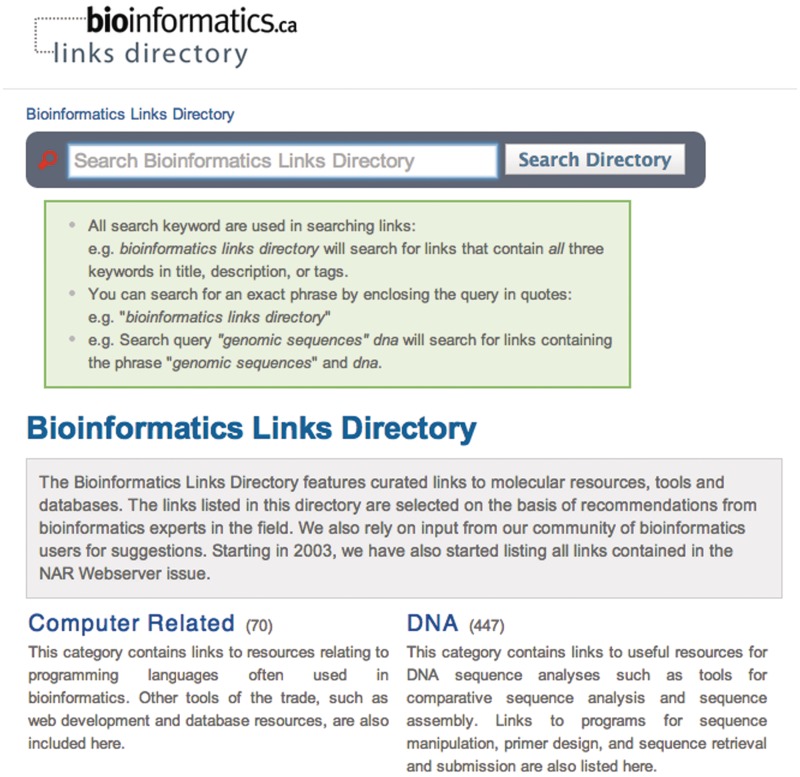
Figure 2.
Searching for links within the Bioinformatics Links Directory may be done through use of keywords in the search box. Suggested search options are provided in the search guide box.
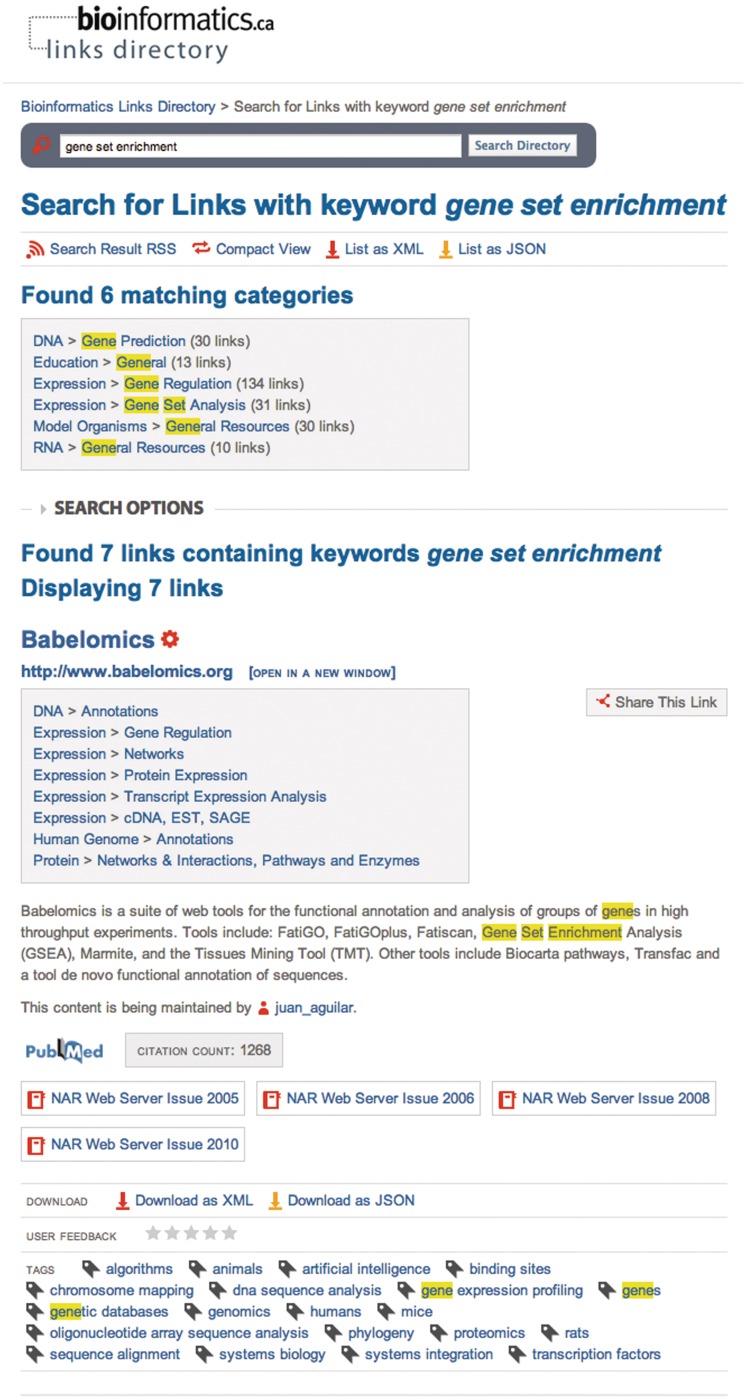
Figure 3.
Keyword searches such as for gene set enrichment return matching categories and links, which contain the requested keywords highlighted in yellow.
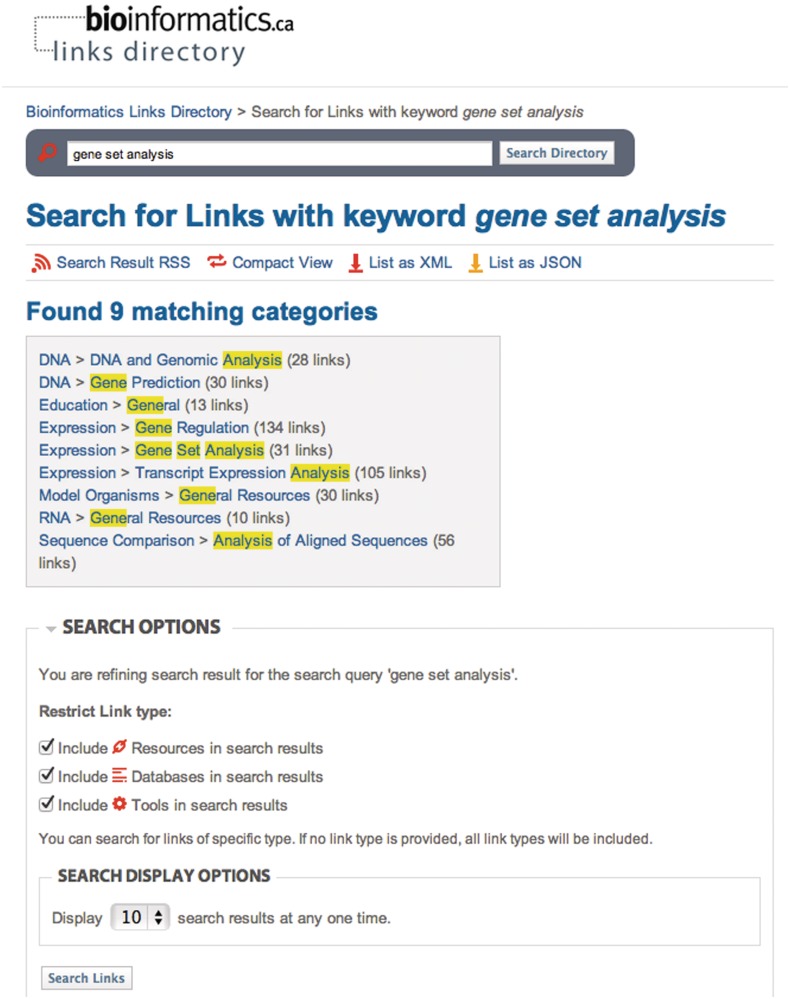
Figure 4.
A returned keyword search may be refined using the Search Options, where returned links may be refined to include or exclude all tools, resources and databases.
Figure 5.
All links within the Bioinformatics Links Directory now contain a per PMID and cumulative Google Scholar citation count for the citations listed for each link, as well as a Google+ and Twitter sharing count.
SHIFTING TRENDS
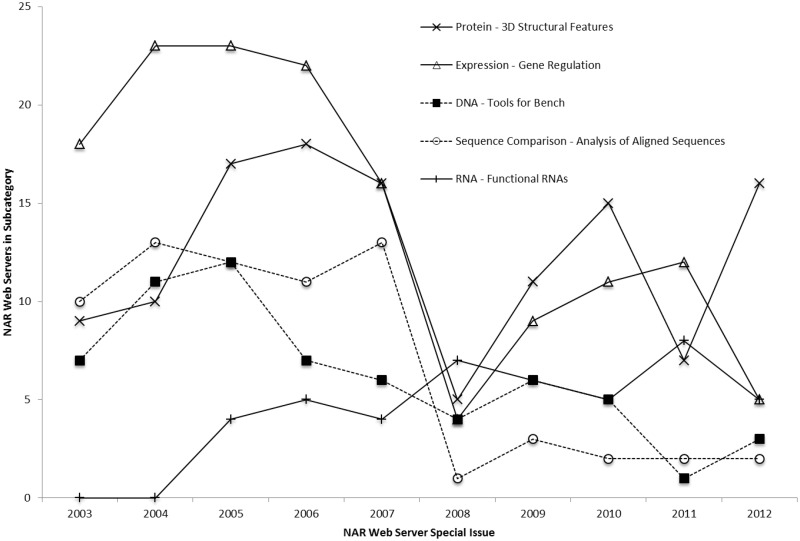
The past decade of NAR Web Server releases has seen significant growth and change in the types of tools, databases and resources being put forth. Although each Web Server issue has seen a steady number of new web servers and updates, the functional utility classification of these web servers has differed greatly over the decade. The Bioinformatics Links Directory in 2006 compiled a graph highlighting the trends in five subcategories of links published by NAR between 2003 and 2006 (3). An update of that graph (Figure 6) shows the exceptional changes in new bioinformatics web servers within these same selected subcategories; Table 1 lists how substantial these changes have been across the entire field of bioinformatics as well (Table 1). Subcategories with active contributions of new web servers in 2006 differ from the subcategories with active contributions of new web servers in 2012 (Figure 6 and Table 1). For example subcategories such as ‘DNA: Tools for the Bench’ subcategory, which contains new web servers for conducting generic DNA analyses such as primer design, exhibited strong growth over 2003–2006 but shows very little growth in 2012 as tools in this category remain relevant over time (Figure 6). In contrast, other subcategories such as Protein: 3D Structural Features have remained active in web server development over time, as the introduction of better technologies and algorithms has permitted a deeper appreciation of proteins and their structural features in 3D (Figure 6).
Figure 6.
Number of NAR Web Servers from selected subcategories of the Bioinformatics Links Directory (http://bioinformatics.ca/links_directory/). The number of NAR Web Servers published from 2003–2012 for five selected subcategories of the Bioinformatics Links Directory: Protein: 3D Structural Features; Expression: Gene Regulation; DNA: Tools for the Bench; Sequence Comparison: Analysis of Aligned Sequences and RNA: Functional RNAs is shown.
Table 1.
Historical summary of the number of resources, databases and web server tools listed in each subcategory of the Bioinformatics Links Directory between 2005 and 2012
| Name | 2005 Total | 2006 Total | 2007 Total | 2008 Total | 2009 Total | 2010 Total | 2011 |
2012a |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Resources | Databases | Tools | Total | Resources | Databases | Tools | Total | |||||||
| Computer related | ||||||||||||||
| Bio-* programming tools | 20 | 20 | 20 | 20 | 20 | 18 | 6 | 12 | 18 | 6 | 12 | 18 | ||
| C/C++ | 3 | 3 | 3 | 3 | 3 | 2 | 1 | 1 | 2 | 1 | 1 | 2 | ||
| Databases | 2 | 2 | 2 | 2 | 3 | 5 | 1 | 5 | 6 | 1 | 5 | 6 | ||
| Java | 4 | 4 | 4 | 4 | 4 | 4 | 3 | 1 | 4 | 3 | 1 | 4 | ||
| Linux/Unix | 12 | 12 | 11 | 11 | 11 | 10 | 5 | 1 | 4 | 10 | 5 | 1 | 4 | 10 |
| PERL | 5 | 5 | 5 | 5 | 5 | 5 | 4 | 1 | 5 | 4 | 1 | 5 | ||
| PHP | 3 | 3 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | ||||
| Statistics | 9 | 9 | 9 | 9 | 9 | 9 | 2 | 7 | 9 | 2 | 7 | 9 | ||
| Web development | 6 | 6 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | ||||
| Web services | 5 | 6 | 7 | 7 | 10 | 17 | 3 | 16 | 19 | 3 | 15 | 18 | ||
| Workflows | 3 | 1 | 5 | 6 | 1 | 7 | 8 | |||||||
| DNA | ||||||||||||||
| Annotations | 29 | 38 | 56 | 57 | 62 | 71 | 1 | 7 | 65 | 73 | 1 | 6 | 59 | 66 |
| Databases | 56 | 56 | 54 | 2 | 56 | |||||||||
| DNA and genomic analysis | 17 | 19 | 20 | 23 | 24 | 25 | 5 | 1 | 22 | 28 | 5 | 1 | 26 | 32 |
| Gene prediction | 17 | 32 | 33 | 34 | 37 | 37 | 1 | 1 | 35 | 37 | 1 | 1 | 29 | 31 |
| Mapping and assembly | 12 | 14 | 15 | 15 | 17 | 20 | 3 | 2 | 16 | 21 | 3 | 2 | 11 | 16 |
| Phylogeny reconstruction | 24 | 37 | 43 | 46 | 49 | 53 | 4 | 4 | 45 | 53 | 4 | 4 | 41 | 49 |
| Sequence polymorphisms | 22 | 32 | 39 | 41 | 42 | 44 | 1 | 8 | 39 | 48 | 1 | 7 | 39 | 47 |
| Sequence retrieval and submission | 23 | 26 | 30 | 32 | 32 | 30 | 1 | 6 | 23 | 30 | 1 | 7 | 20 | 28 |
| Structure and sequence feature detection | 71 | 118 | 142 | 145 | 150 | 161 | 3 | 10 | 150 | 163 | 3 | 9 | 137 | 149 |
| Tools for the bench | 44 | 55 | 63 | 65 | 71 | 76 | 2 | 7 | 67 | 76 | 2 | 4 | 60 | 66 |
| Education | ||||||||||||||
| Bioinformatics-related news sources | 9 | 9 | 9 | 9 | 9 | 9 | 9 | 9 | 7 | 7 | ||||
| Community | 19 | 19 | 24 | 23 | 23 | 18 | 17 | 2 | 19 | 17 | 1 | 1 | 19 | |
| Courses, programs and workshops | 5 | 5 | 5 | 5 | 5 | 4 | 3 | 3 | 1 | 1 | ||||
| Directories and portals | 12 | 15 | 15 | 15 | 15 | 18 | 12 | 6 | 18 | 12 | 1 | 5 | 18 | |
| General | 15 | 15 | 14 | 14 | 14 | 14 | 13 | 1 | 14 | 12 | 1 | 13 | ||
| Tutorials and directed learning resources | 9 | 9 | 9 | 9 | 9 | 10 | 10 | 10 | 9 | 9 | ||||
| Expression | ||||||||||||||
| cDNA, EST, SAGE | 24 | 29 | 36 | 44 | 48 | 49 | 1 | 10 | 38 | 49 | 1 | 8 | 29 | 38 |
| Databases | 21 | 21 | 20 | 3 | 20 | |||||||||
| Gene regulation | 59 | 96 | 119 | 120 | 128 | 138 | 3 | 16 | 127 | 146 | 3 | 16 | 119 | 138 |
| Gene set analysis | 11 | 24 | 1 | 29 | 30 | 1 | 32 | 33 | ||||||
| Networks | 8 | 12 | 16 | 2 | 15 | 17 | 1 | 21 | 22 | |||||
| Protein expression | 7 | 8 | 9 | 17 | 22 | 23 | 1 | 3 | 18 | 22 | 1 | 2 | 18 | 21 |
| Splicing | 9 | 16 | 19 | 19 | 21 | 22 | 3 | 17 | 20 | 3 | 16 | 19 | ||
| Transcript expression and microarrays | 54 | 75 | 89 | 101 | 108 | 121 | 5 | 10 | 107 | 122 | 5 | 9 | 96 | 110 |
| Human genome | ||||||||||||||
| Annotations | 22 | 31 | 37 | 38 | 39 | 46 | 2 | 5 | 39 | 46 | 2 | 4 | 35 | 41 |
| Databases | 31 | 1 | 32 | 30 | 2 | 32 | ||||||||
| Ethics | 7 | 7 | 8 | 8 | 8 | 6 | 5 | 1 | 6 | 4 | 1 | 5 | ||
| Genomics | 4 | 4 | 3 | 3 | 10 | 19 | 3 | 2 | 15 | 20 | 3 | 2 | 16 | 21 |
| Health and disease | 11 | 14 | 19 | 23 | 27 | 29 | 1 | 7 | 23 | 31 | 1 | 7 | 24 | 32 |
| Other resources | 22 | 25 | 29 | 29 | 29 | 31 | 9 | 4 | 17 | 30 | 9 | 4 | 16 | 29 |
| Sequence polymorphisms | 17 | 25 | 33 | 36 | 38 | 46 | 2 | 4 | 43 | 49 | 2 | 4 | 44 | 50 |
| Literature | ||||||||||||||
| Databases | 7 | 7 | 7 | 7 | ||||||||||
| Goldmines | 5 | 6 | 6 | 6 | 6 | 5 | 1 | 4 | 5 | 1 | 4 | 5 | ||
| Open access resources | 2 | 2 | 2 | 2 | 3 | 4 | 1 | 3 | 4 | 1 | 3 | 4 | ||
| Search tools | 6 | 10 | 12 | 12 | 13 | 14 | 6 | 8 | 14 | 6 | 6 | 12 | ||
| Text mining | 7 | 11 | 15 | 22 | 30 | 31 | 4 | 30 | 34 | 4 | 35 | 39 | ||
| Model organisms | ||||||||||||||
| Databases | 95 | 95 | 94 | 1 | 94 | |||||||||
| Fish | 11 | 11 | 11 | 11 | 11 | 11 | 5 | 6 | 11 | 4 | 5 | 9 | ||
| Fly | 12 | 16 | 17 | 17 | 17 | 21 | 2 | 8 | 13 | 23 | 1 | 7 | 16 | 24 |
| General resources | 20 | 23 | 27 | 28 | 29 | 32 | 3 | 12 | 16 | 31 | 3 | 11 | 16 | 30 |
| Microbes | 19 | 31 | 38 | 45 | 53 | 60 | 3 | 12 | 49 | 64 | 2 | 9 | 51 | 62 |
| Mouse and rat | 28 | 32 | 35 | 35 | 36 | 42 | 4 | 17 | 21 | 42 | 4 | 11 | 24 | 39 |
| Other organisms | 13 | 18 | 21 | 21 | 21 | 22 | 3 | 6 | 13 | 22 | 3 | 4 | 13 | 20 |
| Other vertebrates | 10 | 10 | 10 | 10 | 10 | 11 | 1 | 3 | 7 | 11 | 1 | 3 | 7 | 11 |
| Plants | 11 | 16 | 19 | 21 | 25 | 28 | 2 | 10 | 18 | 30 | 2 | 9 | 20 | 31 |
| Worm | 9 | 9 | 9 | 9 | 9 | 10 | 1 | 4 | 6 | 11 | 1 | 4 | 8 | 13 |
| Yeast | 11 | 15 | 18 | 18 | 18 | 21 | 2 | 9 | 10 | 21 | 2 | 7 | 12 | 21 |
| Other molecules | ||||||||||||||
| Carbohydrates | 5 | 6 | 6 | 6 | 6 | 7 | 7 | 7 | 5 | 5 | ||||
| Compounds | 2 | 6 | 12 | 1 | 14 | 15 | 1 | 15 | 16 | |||||
| Databases | 26 | 26 | 25 | 1 | 26 | |||||||||
| Enzymes | 2 | |||||||||||||
| Metabolites | 3 | 4 | 7 | 12 | 1 | 12 | 13 | 1 | 13 | 14 | ||||
| Peptides | 1 | 1 | ||||||||||||
| Small molecules | 1 | 3 | 6 | 6 | 9 | 13 | 2 | 13 | 15 | 2 | 15 | 17 | ||
| Protein | ||||||||||||||
| 2D Structure prediction | 29 | 51 | 58 | 60 | 63 | 65 | 1 | 65 | 66 | 56 | 56 | |||
| 3D Structural features | 53 | 70 | 75 | 85 | 100 | 1 | 2 | 103 | 106 | 1 | 2 | 107 | 110 | |
| 3D Structure comparison | 78 | 35 | 45 | 50 | 59 | 71 | 1 | 6 | 68 | 75 | 1 | 6 | 62 | 69 |
| 3D Structure prediction | 48 | 59 | 60 | 70 | 83 | 4 | 1 | 85 | 90 | 2 | 1 | 83 | 86 | |
| 3D Structure retrieval, viewing | 28 | 45 | 51 | 52 | 56 | 58 | 4 | 7 | 47 | 58 | 3 | 8 | 42 | 53 |
| Annotation and function | 26 | 35 | 44 | 47 | 53 | 57 | 5 | 57 | 62 | 5 | 56 | 61 | ||
| Biochemical features | 25 | 37 | 40 | 41 | 46 | 46 | 2 | 2 | 43 | 47 | 2 | 2 | 41 | 45 |
| Databases | 116 | 1 | 117 | 115 | 2 | 117 | ||||||||
| Do-it-all tools for protein | 6 | 8 | 8 | 13 | 14 | 15 | 3 | 12 | 15 | 3 | 11 | 14 | ||
| Domains and motifs | 50 | 86 | 112 | 115 | 121 | 124 | 14 | 115 | 129 | 14 | 109 | 123 | ||
| Identification, presentation and format | 12 | 13 | 14 | 14 | 14 | 14 | 1 | 13 | 14 | 1 | 14 | 15 | ||
| Networks & Interactions, pathways, enzymes | 48 | 66 | 88 | 94 | 107 | 125 | 3 | 21 | 113 | 137 | 2 | 19 | 113 | 134 |
| Localization and targeting | 19 | 30 | 38 | 38 | 39 | 41 | 3 | 39 | 42 | 3 | 34 | 37 | ||
| Molecular dynamics and docking | 19 | 21 | 27 | 34 | 40 | 1 | 44 | 45 | 1 | 48 | 49 | |||
| Phylogeny reconstruction | 28 | 36 | 44 | 45 | 53 | 54 | 4 | 5 | 47 | 56 | 4 | 4 | 48 | 56 |
| Protein expression | 7 | 8 | 8 | 8 | 10 | 10 | 1 | 2 | 7 | 10 | 1 | 2 | 8 | 11 |
| Proteomics | 21 | 25 | 27 | 33 | 37 | 39 | 3 | 4 | 32 | 39 | 3 | 4 | 26 | 33 |
| Sequence comparison | 7 | 14 | 17 | 18 | 18 | 18 | 18 | |||||||
| Sequence data | 7 | 7 | 8 | 9 | 10 | 10 | 1 | 6 | 3 | 10 | 1 | 5 | 3 | 9 |
| Sequence features | 14 | 25 | 31 | 33 | 38 | 46 | 2 | 2 | 46 | 50 | 2 | 3 | 47 | 52 |
| Sequence retrieval | 21 | 27 | 29 | 29 | 31 | 29 | 1 | 6 | 21 | 28 | 1 | 7 | 20 | 28 |
| RNA | ||||||||||||||
| Databases | 0 | 24 | 1 | 25 | 23 | 2 | 25 | |||||||
| Functional RNAs | 9 | 14 | 19 | 26 | 32 | 37 | 0 | 5 | 39 | 44 | 5 | 41 | 46 | |
| General resources | 8 | 10 | 10 | 10 | 10 | 11 | 4 | 3 | 4 | 11 | 5 | 2 | 4 | 11 |
| Motifs | 9 | 19 | 21 | 22 | 23 | 25 | 0 | 1 | 26 | 27 | 1 | 25 | 26 | |
| Sequence retrieval | 11 | 11 | 10 | 11 | 11 | 9 | 0 | 2 | 7 | 9 | 2 | 7 | 9 | |
| Structure prediction, visualization and design | 24 | 38 | 47 | 54 | 58 | 62 | 0 | 3 | 65 | 68 | 3 | 64 | 67 | |
| Sequence comparison | ||||||||||||||
| Alignment editing and visualization | 14 | 20 | 21 | 21 | 23 | 25 | 0 | 0 | 25 | 25 | 23 | 23 | ||
| Analysis of aligned sequences | 27 | 43 | 59 | 60 | 62 | 64 | 0 | 0 | 65 | 65 | 1 | 57 | 58 | |
| Comparative genomics | 19 | 26 | 33 | 35 | 37 | 48 | 2 | 7 | 40 | 49 | 2 | 6 | 33 | 41 |
| Multiple sequence alignments | 19 | 38 | 50 | 56 | 57 | 65 | 1 | 0 | 67 | 68 | 1 | 1 | 62 | 64 |
| Other alignment tools | 6 | 11 | 11 | 11 | 11 | 12 | 0 | 0 | 13 | 13 | 12 | 12 | ||
| Pairwise sequence alignments | 11 | 22 | 23 | 26 | 33 | 35 | 1 | 0 | 34 | 35 | 1 | 30 | 31 | |
| Similarity searching | 13 | 31 | 47 | 47 | 49 | 50 | 1 | 2 | 49 | 52 | 1 | 3 | 45 | 49 |
| Total unique resources, databases and tools in Bioinformatics Links Directory | 700+ | 1000+ | 1100+ | 1200+ | 1400 | 1500 | 144 | 480 | 1250 | 1874 | 134 | 455 | 1205 | 1794 |
aA complete listing of all URLs listed in the Nucleic Acids Research 2012 Web Server Issue can be accessed online at: http://bioinformatics.ca/links_directory/narweb2012.
Indeed, many of these trends in subcategory activity reflect the introduction of a new research technology and/or an underlying research need. As originally discussed in 2009, yet still evident throughout Table 1, there are observable sharp spikes in the number of informatics tools available in a given biological area or specific to a particular task (6). Although between 2006 and 2007, there was a spike in ‘DNA: Structure and Sequence Feature Detection’, ‘Expression: Gene Regulation’ and ‘Expression: Transcript Expression and Microarrays’ most likely in response to the pervasive use of expression platforms and transcript analyses at that time, current spikes in ‘Human Genome: Genomics’, ‘Sequence Comparison: Comparative Genomics’ and ‘Sequence Comparison: Pairwise Sequence Alignments’ between 2010 and 2012 may be reflective of the introduction of next-generation sequencing technologies. Table 1 highlights all the trends between 2005 and 2012 across all subcategories of the Bioinformatics Links Directory (Table 1) (individual calendar year data are not available for 2003–2005 as the Bioinformatics Links Directory collaboration with NAR began in 2005 for all data before that). Of note are the aggressive coverage of databases, which began in 2011 and the need for additional subcategories such as ‘Other Molecules: Enzymes and Peptides’ in response to the growing diversity and depth in the field.
CONSTANT GROWTH
The number of bioinformatics links produced by the community has nonetheless continued to accumulate over time. When first released in 2002 under Bioinformatics.ca, there were only 385 links in the database. By 2005, when the Bioinformatics Links Directory began its collaboration with NAR, the Directory already contained 700+ web servers and resources (2). The Bioinformatics Links Directory has continued to grow (and diminish in size as web servers become extinct) with each successive year (Table 1). The Bioinformatics Links Directory now contains more than 1200 unique web servers. This year's NAR Web Server Issue introduces an additional 90 web servers and 12 server updates (Table 1). The complete listing of URLs cited in the 2012 Web Server Issue can be accessed online at the Nucleic Acids Research website, http://nar.oxfordjournals.org/, and at http://bioinformatics.ca/links_directory/narweb2012/.
Growth has also resulted in change. In 2011, the Bioinformatics Links Directory introduced all the databases published by Nucleic Acids Research over their 2010–2011 window and completed a major reorganization of its existing links (8). Links were reclassified into one of three types reflecting the growing variety of bioinformatics materials being generated to support research: (i) Resource—a static resource whose intention is to convey bioinformatic information; (ii) Tool—a bioinformatic web server or downloadable software tool that can query, analyze, extract or modify input data and (iii) Database—a biological data store that can be queried. Under this new schema and with this expanded content, the Bioinformatics Links Directory now contains 1794 links comprising 455 databases, 134 resources and 1205 web server tools (Table 1). The Bioinformatics Links Directory has also initiated active curation of its content, removing dead content and correcting content errors, which has resulted in more accurate although occasionally smaller counts for 2012 (Table 1). Over time, the Bioinformatics Links Directory is expected to continue its growth and evolution in keeping with the current pace of research.
FUTURE CHALLENGES
With such a vast number of bioinformatics tools available for biological research, now more than ever, there is a need for a well-annotated and user-friendly ‘resourceome’ (1). In 2011, the Bioinformatics Links Directory introduced features to facilitate peer review of its links and link ownership (8). The application of Google +1 and other social media features, such as Twitter to the Bioinformatics Links Directory, provides users with the opportunity to share useful links; registered users may also post comments and review a link’s usefulness or start a discussion forum. Along with enhanced search capabilities and MeSH term and keyword tagging, such a peer-review system allows users to rapidly narrow down the links most applicable to their research need. Other features added provide link owners with the capacity to edit, manage and communicate on their own link without requiring input or support from the Bioinformatics Links Directory (8).
Although necessary to remain current and to advance the utility of the Bioinformatics Links Directory, these improvements will only prove useful if driven by the community. As a community-driven repository, everyone in the research or bioinformatics community has the opportunity to help make the collection better and more meaningful. Anyone may (i) suggest a link through links@bioinformatics.ca or through the ‘Suggest a URL’ page on the Bioinformatics Links Directory; (ii) register for a free account and submit their own tool or database; (iii) rate a tool, resource or database; (iv) register for a free account and submit their review of a tool, resource or database; (v) enrich a tool or database with tutorials or user-oriented content; (vi) initiate a discussion forum on a problem related to the use of any given tool or database or (vii) share a useful tool, resource or database with their colleagues. After a decade of output from the NAR Web Server issues, it will be interesting to monitor the development, maturity and impact of these bioinformatics web servers and databases over the next decade.
FUNDING
Ontario Institute for Cancer Research and the government of Ontario, and from Genome Canada and the Ontario Genome Institute though a platform award to the The Center of Applied Genomics (TCAG) at the Hospital for Sick Children (SickKids). Funding for open access charge: Waived by Oxford University Press.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The Bioinformatics Links Directory is a community resource built on a commitment to the spirit of open access under the Attribution-Share Alike 2.5 Canada (CC BY-SA 2.5) license. The authors acknowledge the efforts of Nucleic Acids Research and the researchers and developers worldwide who invest considerable effort into ensuring that their research is freely accessible to all. In particular, the authors acknowledge all the contributors to the Bioinformatics Links Directory for their valuable input and suggestions for improvements to the directory; these individuals are listed on the Acknowledgements page at http://bioinformatics.ca/links_directory/acknowledgements/. In particular, they recognize the continued support of the OICR Web Development group, especially Joseph T. Yamada.
REFERENCES
- 1.Cannata N, Merelli E, Altman RB. Time to organize the bioinformatics resourceome. PLoS Comput. Biol. 2005;1:e76. doi: 10.1371/journal.pcbi.0010076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fox JA, Butland SL, McMillan S, Campbell G, Ouellette BF. The Bioinformatics Links Directory: a compilation of molecular biology web servers. Nucleic Acids Res. 2005;33:W3–W24. doi: 10.1093/nar/gki594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fox JA, McMillan S, Ouellette BF. A compilation of molecular biology web servers: 2006 update on the Bioinformatics Links Directory. Nucleic Acids Res. 2006;34:W3–W5. doi: 10.1093/nar/gkl379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fox JA, McMillan S, Ouellette BF. Conducting research on the web: 2007 update for the bioinformatics links directory. Nucleic Acids Res. 2007;35:W3–W5. doi: 10.1093/nar/gkm459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brazas MD, Fox JA, Brown T, McMillan S, Ouellette BF. Keeping pace with the data: 2008 update on the Bioinformatics Links Directory. Nucleic Acids Res. 2008;36:W2–W4. doi: 10.1093/nar/gkn399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brazas MD, Yamada JT, Ouellette BF. Evolution in bioinformatic resources: 2009 update on the Bioinformatics Links Directory. Nucleic Acids Res. 2009;37:W3–W5. doi: 10.1093/nar/gkp531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brazas MD, Yamada JT, Ouellette BF. Providing web servers and training in Bioinformatics: 2010 update on the Bioinformatics Links Directory. Nucleic Acids Res. 2010;38:W3–W6. doi: 10.1093/nar/gkq553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brazas MD, Yim DS, Yamada JT, Ouellette BF. The 2011 Bioinformatics Links Directory update: more resources, tools and databases and features to empower the bioinformatics community. Nucleic Acids Res. 2011;39:W3–W7. doi: 10.1093/nar/gkr514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Google Scholar. http://scholar.google.ca/ (10 June 2012, date last accessed)
- 10. Google+. http://www.google.com/+1/button/ (10 June 2012, date last accessed)