Abstract
Binding hot spots, protein sites with high-binding affinity, can be identified using X-ray crystallography or NMR by screening libraries of small organic molecules that tend to cluster at such regions. FTMAP, a direct computational analog of the experimental screening approaches, globally samples the surface of a target protein using small organic molecules as probes, finds favorable positions, clusters the conformations and ranks the clusters on the basis of the average energy. The regions that bind several probe clusters predict the binding hot spots, in good agreement with experimental results. Small molecules discovered by fragment-based approaches to drug design also bind at the hot spot regions. To identify such molecules and their most likely bound positions, we extend the functionality of FTMAP (http://ftmap.bu.edu/param) to accept any small molecule as an additional probe. In its updated form, FTMAP identifies the hot spots based on a standard set of probes, and for each additional probe shows representative structures of nearby low energy clusters. This approach helps to predict bound poses of the user-selected molecules, detects if a compound is not likely to bind in the hot spot region, and provides input for the design of larger ligands.
INTRODUCTION
Hot spots are locations on the protein surface that contribute significantly to the ligand binding free energy, and are important targets in many biological applications including rational drug design. The locations of these hot spots can be identified by screening a protein of interest against libraries of small organic molecules using NMR spectroscopy (1,2) or X-ray crystallography (3). The congregation of many types of small organic molecules in selected locations identifies the binding hot spots on the protein surface. The biophysical basis of this phenomenon is not fully understood, but many studies had substantiated this observation time and again (1–3). Fesik et al. (2) demonstrated the propensity of hot spots to bind many types of small organic molecules using NMR spectroscopy-based screening. The multiple solvent crystal structures (MSCS) method, based on X-ray crystallography, superimposes the structures of the target protein solved in 8–10 types of organic solutions to find clusters of small molecules (3).
The identification of hot spots using biophysical methods such as NMR spectroscopy and MSCS is costly, time-consuming and is limited by physical constraints such as the solubility of the small organic molecules. FTMAP is a computational analog of these experimental approaches (4–9). The method places molecular probes—small organic molecules that vary in size, shape and polarity—on a dense grid around the protein, and finds favorable positions using first an empirical energy function and then the CHARMM potential with a continuum electrostatics term. A number of low energy conformations are clustered and the clusters ranked on the basis of the average energy. The regions that bind several probe clusters are the predicted hot spots, and the one binding the largest number of probe clusters is considered the main hot spot. It was shown that FTMAP is capable of identifying the binding hot spots using a set of 16 small organic molecules as probes, in good agreement with the results of SAR by NMR and MSCS experiments (4). Aqueous solutions of most of these probes have also been used for soaking protein crystals in the MSCS experiments, which helps to directly compare the observed and predicted positions.
The identification of hot spots plays an important role in fragment-based drug design (FBDD). FBDD generally starts with finding fragment-sized compounds that are highly ligand efficient and can serve as a core moiety for developing high-affinity leads. Such core moieties are most frequently found by soaking protein crystals in mixtures of compounds from a library of fragment-size molecules with functional groups that occur in known drugs. As recently shown, the core fragments always bind within the main hot spot identified by FTMAP. Additional secondary hot spots near the main hot spot show whether the core fragment can be extended and, if so, which directions are best for extension (9). These results have three important applications. First, the information helps to find the bound pose of potential cores, as such molecules always overlap with the main hot spot. In fact, it is frequently difficult to dock small molecules to proteins because they can fit into a number of pockets, in addition to the functional binding site, and current scoring functions provide limited accuracy for the elimination of false-positive positions. It was recently shown that searching for maximum correlation with the density of probes obtained by the mapping helps to locate the most likely poses of bound ligands. Second, if a small molecule has no docked position in the hot spot region then it is not likely to serve as a potential core. Third, the position and orientation of the fragment-sized molecules in the main and secondary hot spots provide input for the design of larger ligands that include several of the functional groups occurring in different fragments.
In its earlier implementation, the FTMAP server could use only the pre-defined set of 16 molecules as probes (4). In view of the above discussion, it is of substantial interest to determine the distribution of bound poses for a variety of fragment-sized candidate molecules around the hot spots, i.e. adding such molecules to the standard probe set. Extending FTMAP to arbitrary probes we face two problems. First, the 16 molecules in our standard set have molecular weights below 100 and have no rotational degrees of freedom, whereas the libraries of fragment-sized compounds used for screening in FBDD usually consists of compounds with molecular weight in the range of 150–250 and with one or two rotatable bonds. Since the first stage of mapping is a rigid body global search, it is necessary to generate the set of the most likely rotamers. The extended FTMAP server accepts user-supplied small molecules as SMILES strings, and generates conformers using the program Confab (10) to be used alongside the 16 standard types of small molecules. Second, mapping needs a substantial number of parameters, both for the grid search and for the minimization by CHARMM. Parameters for the additional probes are generated by a variety of computational chemistry programs including ANTECHAMBER (11) based on the general AMBER force field (GAFF) (12), and General Atomic and Molecular and Electronic Structure Systems (GAMESS)(13). The charge model Austin Model 1 bond charge correction (AM1-BCC) (14) had been chosen to calculate atomic charges because the good quality of the charge assignments is similar to those computed using an ab initio scheme (15) but incurs much lower computational costs. The server can also be used for generating parameters only, i.e. without running an FTMAP analysis. The generated topology and parameter files can consequently be used in any application that requires CHARMM (16) file formats. The run time for mapping a protein is about 2 h when using only 16 types of probes, but can be longer if the user submits many additional molecules, or if the target protein is very large.
RESULTS
Protein mapping
FTMAP globally samples the surface of a protein on a dense grid using 16 types of small molecules as probes (ethanol, isopropanol, isobutanol, acetone, acetaldehyde, dimethyl ether, cyclohexane, ethane, acetonitrile, urea, methylamine, phenol, benzaldehyde, benzene, acetamide and N,N-dimethylformamide). FTMAP performs four steps as follows (4).
The rotational/translational space of each probe is systematically sampled on a grid around the fixed protein, consisting of 0.8 Å translations and of 500 rotations at each location. The energy function includes a stepwise approximation of the van der Waals energy with attractive and repulsive contributions, and an electrostatics/solvation term based on Poisson–Boltzmann continuum model (17) with the dielectric constants of ε = 4 and ε = 80 for the protein and the solvent, respectively. The 2000 best poses for each probe are retained for further processing.
The 2000 complexes are refined by off-grid energy minimization during which the protein atoms are held fixed while the atoms of the probe molecules are free to move. The energy function includes the bonded and van der Waals terms of the CHARMM potential and an electrostatics/solvation term based on the analytical continuum electrostatic (ACE) model as implemented in CHARMM (16).
The minimized probe conformations are grouped into clusters using a simple greedy algorithm and a 4 Å RMSD clustering radius. Clusters with <10 members are excluded from consideration. The retained clusters are ranked on the basis of their Boltzmann averaged energies. Six clusters with the lowest average energies are retained for each probe.
To determine the hot spots, FTMAP finds consensus sites (CSs), i.e. regions on the protein where clusters of different probes overlap. Therefore, the probe clusters are clustered again using the distance between the centers of mass of the cluster centers as the distance measure and 4 Å as the clustering radius. The CSs are ranked based on the number of their clusters, with duplicate clusters of the same type also considered in the count. The largest CS defines the most important hot spot, with smaller CSs identifying secondary hot spots that generally also contribute to ligand binding.
In this implementation, identical to the methodology used by Brenke et al. (4), the CSs are defined only by the clusters of the 16 probe types in the standard set, as these molecules provide reliable and stable hot spot information. The user-supplied small molecules are not used for hot spot identification, but the centers of the lowest energy clusters of these molecules within a radius of 4.0 Å from the center of the CSs are shown as additional results. For each conformer of each additional probe, the results online show only the lowest energy representative, but the top six lowest energy representatives can be found in the downloadable PyMol session. Some of the user-selected molecules may have no low energy clusters near any of the hot spots, which implies that they are not likely to bind in the hot spot region identified by FTMAP.
Parameterization of small molecules for mapping
The updated FTMAP server accepts user-supplied small molecules in the SMILES format. On the FTMAP main submission page, the user can either directly transfer the generated parameters to mapping, or download the files for inspection and editing and then upload them to be used in the mapping. The examples on the website provide step-by-step instructions. The user can also choose to generate topology and parameter files for the selected compounds in CHARMM file format without mapping any protein. The generation of parameters is based on a number of frequently used programs. The computational chemistry toolset RDKIT (RDKIT: Open-source chemoinformatics; http://www.rdkit.org) is used to generate 3D coordinates based on the submitted SMILES strings, and the program OpenBabel (18) performs all required chemical file format translations. The tool Confab (10) is then used to generate multiple conformations of the small molecules. Confab's methods utilize the MMFF94 force field to calculate the energy as it rotates single bonds between heavy atoms (excluding rings) if the contributing atoms connect to at least one additional heavy atom. The program then calculates the heavy atom RMSD to return only structures that differ by at least 0.5 Å. Due to computational limitations, only structures with fewer than 99 generated conformations (or about three rotatable bonds) are parameterized, but this constraint is generally not a problem when considering compounds used as potential starting points in FBDD.
Subsequent to the generation of 3D coordinates, the quantum chemistry system GAMESS (18) is used to compute AM1 atomic charges; the information is then piped through molecule manipulation scripts from the Sarnoff Corporation (http://charles.karney.info/b2d-scripts/) using the molecular mechanics suite ANTECHAMBER (11) to perform BCCs and to generate the final AM1-BCC atomic charges. ANTECHAMBER generates GAFF-based (12) topology files and parameter files in the CHARMM (20) format. The GAFF (12) force field was constructed based on a number of sources, including parameters from the AMBER force field, crystal structures and ab initio optimizations (MP2/6-31G*) of nearly 2000 model molecules. Since the development of GAFF involved small organic compounds, the resulting parameters are generally transferable to a broad range of such compounds. GAFF is a complete force field, i.e. parameters are either available for all atom types or can be computed using empirical rules (12). Consequently, GAFF is capable of supplying parameters for most organic molecules of pharmaceutical significance composed of the atom types C, N, O, S, P, H and halogens, and is suitable for the modeling of molecules used in drug design.
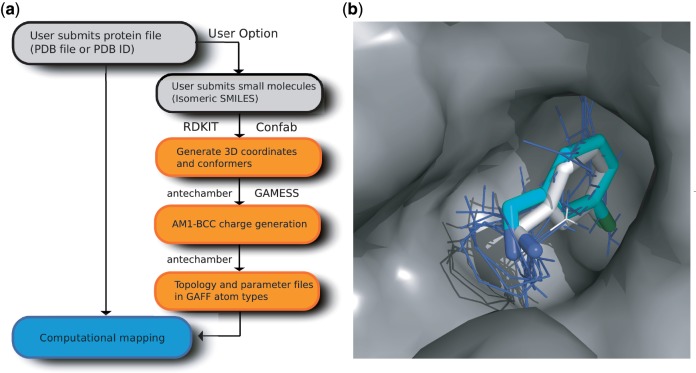
The calculation of partial charges using the AM1-BCC model consists of two steps. First, AM1 atomic charges are generated, followed by BCCs to obtain charges similar to the RESP (19) model. The general shortcoming of semi-empirical charge models is the poor transferability of parameters if the ligand of interest substantially differs from the molecules in the empirical data set used for constructing the parameters. To ensure transferability, Jakalian et al. (14) generated AM1 charges for 2755 molecules using MOPAC 6.0, and then computed BCC parameters by fitting against electrostatic potentials (ESPs) of these molecules, obtained by high-quality HF/6-31G* quantum mechanical calculations. The atom and bond types used to define these BCCs fully sample the atom and bond types from the Merck Index (10 000 molecules) and from the National Cancer Institute Database (250 000 molecules). Therefore, the AM1-BCC model can rapidly generate atomic charges of similar quality to the RESP (19) model for many small organic molecules. Figure 1a shows the schematic diagram of the updated FTMAP server.
Figure 1.
Extended FTMAP server. (a) Schematic diagram of the updated implementation and the generation of force field parameters for user-supplied small molecules; programs used are indicated next to the appropriate boxes. (b) Mapping of an unbound structure of thrombin (PDB ID: 1HXF) using the small molecule C2A from a ligand-bound thrombin structure (PDB ID: 2C8Z) as a user-supplied additional probe. The lowest energy cluster of C2A (with the cluster shown as cyan sticks) overlaps the main consensus site (blue lines) from the mapping of the unbound thrombin structure using the standard probe set, and has an almost identical pose to the ligand from the bound structure (white sticks). Nitrogen and chlorine atoms are coloured blue and green, respectively.
Using the server
The updated server is accessible online (http://ftmap.bu.edu/param) and has been tested using Google Chrome and Mozilla Firefox. The use of the server is free and open to all users, and there is no login requirement. Users can go through a test application under the ‘Examples’ tab for step-by-step instructions before submitting a job. The interface is minimalistic: as standard input, the user is required to supply either the PDB ID of the target protein or to upload a PDB coordinate file. The user also needs to enter the PDB chain ID to be used in the mapping. Following the input of the PDB ID (or uploading a PDB coordinate file) and chain identifiers, the user can choose either to use only the 16 small molecules for the mapping or to supplement the set with up to 10 additional molecules. These additional compounds are specified by their SMILES strings and formal charges (to the left of the SMILES strings). Isomeric SMILES must be used to specify the stereochemistry. The server generates conformers and compute physical parameters for each user-selected probe. If the user wants to inspect and edit the parameters of these compounds, then the SMILES strings should be submitted using the stand-alone parameterization function of the server by going to the ‘Parameterization’ tab. In addition, as described earlier, the parameterization server can be used to generate topology and parameter files in the CHARMM format, without submitting a protein to FTMAP for analysis.
There are two restrictions on the use of our current FTMAP implementation. First, in order to limit computer time required for the mapping, the user can only submit SMILES strings of small molecules containing three or less rotatable bonds; typically this translates up to 100 conformers per compound. FTMAP is hosted on institutional computing facilities and because of the constraints on resources, excessively demanding computations are not feasible. Second, mapping proteins of 800 or more residues can sometimes fail because of similar constraints.
Case studies
To demonstrate the function of the updated FTMAP server, we show the mapping of thrombin. Results are accessible on the FTMAP homepage under the ‘Examples’ tab by selecting ‘Sample Run’. The unbound structure of thrombin (PDB ID: 1HXF) (19) was mapped using the 16 standard probes and an additional small molecule (HETATM ID: C2A) used in an FBDD campaign and co-crystallized with thrombin (PDB ID: 2C8Z) (20). FTMAP identifies the important hot spots based on the consensus clusters of the 16 standard probes, and reports the lowest energy cluster representatives of C2A within 4 Å of each of the consensus cluster, using the geometric center distances in the calculations. Figure 1b shows one of the lowest energy poses of C2A generated by computational mapping on top of the most populated consensus cluster. It is interesting to note that this pose is almost identical to the bound pose of C2A, co-crystallized with thrombin (PDB ID: 2C8Z). Although C2A is a weak binder with an IC50 of only 300 µM, FTMAP was capable of detecting the interaction. The pose identified by computational mapping of C2A is particularly interesting, because the chlorophenyl group occupies the S1 site fully and the NH2 group protrudes from the pocket, indicating the possibility of expanding the molecule. Indeed, C2A was subsequently joined with a 12 µM ligand to generate a 220 nM inhibitor (20). Therefore, in this case, computational mapping recapitulated important protein–ligand interactions, and this type of information can be very useful for screening candidate molecules in FBDD.
Our earlier implementation of the FTMAP algorithm (4), using only the 16 standard probes, is still available at http://ftmap.bu.edu/. Under the ‘Examples’ tab, the site shows mapping results for five additional proteins, angiotensin converting enzyme, carbonic anhydrase I, neuraminidase N2, phospholipase A2 and urokinase type plasminogen activator. An unbound structure, downloaded from the Protein Data Bank (21) was mapped for each protein. We have re-mapped the same structures (without any additional ligand) using the new server, and added the results under the ‘Examples’ tab. In each case, a ligand-bound form of the protein is aligned to the unbound form used in the computational mapping in order to show that the important CSs generally overlap with the positions of the bound ligands, and identify subsites of the ligand binding sites. Comparison of the results obtained by the two servers shows some differences, in spite of a qualitative agreement. Specifically, both the location and the ranking of the large hot spots remain unchanged for neuraminidase N2, phospholipase A2 and urokinase type plasminogen activator. For angiotensin converting enzyme, the second largest consensus cluster in the original FTMAP becomes the third one in the updated FTMAP results. For carbonic anhydrase I the second consensus cluster becomes the first (15 clusters in both cases), whereas the top consensus cluster becomes the third (17 clusters in the original and 13 in the updated results). These differences are primarily due to replacing the Poisson–Boltzmann equation solver of CHARMM (16), used in the earlier server, by the much faster APBS 1.3 program (22) in the calculation of the continuum electrostatics term. In addition, we perform the calculations in double precision. All other elements of the FTMAP algorithm remain unchanged.
CONCLUSIONS
The protein mapping algorithm FTMAP (4) is a computational analog of experimental screening approaches, based on NMR (1,2) or X-ray crystallography (3), to the identification of binding hot spots of proteins. The demonstrated robustness of mapping contrasts the uncertainty of finding bound poses of small ligands by traditional docking methods. FTMAP was originally implemented as a server that used only a standard set of 16 small molecules as probes. While this probe set is sufficient for the reliable identification of hot spots (4), it was shown that finding the distribution of further small molecules around the hot spot region can be very useful for FBDD (9). The extension of FTMAP described here enables the user to submit arbitrary small molecules for mapping. FTMAP identifies the hot spots using the standard probes, and for each additional probe provides representative poses of the lowest energy clusters located close to the hot spots. These results help to find bound poses for the user-specified molecules, show if a compound is not likely to bind in the hot spot region and provide input for the design of larger ligands (9).
FUNDING
National Institute of General Medical Sciences [GM064700]. Funding for open access charge: National Institutes of Health.
Conflict of interest statement. None declared.
REFERENCES
- 1.Liepinsh E, Otting G. Organic solvents identify specific ligand binding sites on protein surfaces. Nat. Biotechnol. 1997;15:264–268. doi: 10.1038/nbt0397-264. [DOI] [PubMed] [Google Scholar]
- 2.Hajduk PJ, Huth JR, Fesik SW. Druggability indices for protein targets derived from NMR-based screening data. J. Med. Chem. 2005;48:2518–2525. doi: 10.1021/jm049131r. [DOI] [PubMed] [Google Scholar]
- 3.Mattos C, Ringe D. Locating and characterizing binding sites on proteins. Nat. Biotechnol. 1996;14:595–599. doi: 10.1038/nbt0596-595. [DOI] [PubMed] [Google Scholar]
- 4.Brenke R, Kozakov D, Chuang GY, Beglov D, Hall D, Landon MR, Mattos C, Vajda S. Fragment-based identification of druggable ‘hot spots’ of proteins using Fourier domain correlation techniques. Bioinformatics. 2009;25:621–627. doi: 10.1093/bioinformatics/btp036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ngan CH, Beglov D, Rudnitskaya AN, Kozakov D, Waxman DJ, Vajda S. The structural basis of pregnane X receptor binding promiscuity. Biochemistry. 2009;48:11572–11581. doi: 10.1021/bi901578n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chuang GY, Mehra-Chaudhary R, Ngan CH, Zerbe BS, Kozakov D, Vajda S, Beamer LJ. Domain motion and interdomain hot spots in a multidomain enzyme. Protein Sci. 2010;19:1662–1672. doi: 10.1002/pro.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kozakov D, Hall DR, Chuang GY, Cencic R, Brenke R, Grove LE, Beglov D, Pelletier J, Whitty A, Vajda S. Structural conservation of druggable hot spots in protein-protein interfaces. Proc. Natl Acad. Sci. USA. 2011;108:13528–13533. doi: 10.1073/pnas.1101835108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ngan CH, Hall DR, Zerbe B, Grove LE, Kozakov D, Vajda S. FTSite: high accuracy detection of ligand binding sites on unbound protein structures. Bioinformatics. 2012;28:286–287. doi: 10.1093/bioinformatics/btr651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hall DR, Ngan CH, Zerbe BS, Kozakov D, Vajda S. Hot spot analysis for driving the development of hits into leads in fragment-based drug discovery. J. Chem. Inf. Model. 2012;52:199–209. doi: 10.1021/ci200468p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O'Boyle NM, Vandermeersch T, Flynn CJ, Maguire AR, Hutchison GR. Confab: systematic generation of diverse low-energy conformers. J. Cheminform. 2011;3:8. doi: 10.1186/1758-2946-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang J, Wang W, Kollman PA, Case DA. Automatic atom type and bond type perception in molecular mechanical calculations. J. Mol. Graph. Model. 2006;25:247–260. doi: 10.1016/j.jmgm.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 12.Wang J, Wolf RM, Caldwell JW, Kollman PA, Case DA. Development and testing of a general amber force field. J. Comput. Chem. 2004;25:1157–1174. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- 13.Schmidt MW, Baldridge KK, Boatz JA, Elbert ST, Gordon MS, Jensen JH, Koseki S, Matsunaga N, Nguyen KA, Su SJ, et al. General atomic and molecular electronic-structure system. J. Comput. Chem. 1993;14:1347–1363. [Google Scholar]
- 14.Jakalian A, Jack DB, Bayly CI. Fast, efficient generation of high-quality atomic charges. AM1-BCC model: II. Parameterization and validation. J. Comput. Chem. 2002;23:1623–1641. doi: 10.1002/jcc.10128. [DOI] [PubMed] [Google Scholar]
- 15.Bayly CI, Cieplak P, Cornell WD, Kollman PA. A well-behaved electrostatic potential based method using charge restraints for deriving atomic charges: the RESP model. J. Phys. Chem. 1993;97:10269–10280. [Google Scholar]
- 16.Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M. Charmm: a program for macromolecular energy, minimization, and dynamics calculations. J. Comput. Chem. 1983;4:187–217. [Google Scholar]
- 17.Gilson MK, Honig B. Calculation of the total electrostatic energy of a macromolecular system: solvation energies, binding energies, and conformational analysis. Proteins. 1988;4:7–18. doi: 10.1002/prot.340040104. [DOI] [PubMed] [Google Scholar]
- 18.Guha R, Howard MT, Hutchison GR, Murray-Rust P, Rzepa H, Steinbeck C, Wegner J, Willighagen EL. The Blue Obelisk-interoperability in chemical informatics. J. Chem. Inf. Model. 2006;46:991–998. doi: 10.1021/ci050400b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang E, Tulinsky A. The molecular environment of the Na+ binding site of thrombin. Biophys. Chem. 1997;63:185–200. doi: 10.1016/s0301-4622(96)02227-2. [DOI] [PubMed] [Google Scholar]
- 20.Howard N, Abell C, Blakemore W, Chessari G, Congreve M, Howard S, Jhoti H, Murray CW, Seavers LC, van Montfort RL. Application of fragment screening and fragment linking to the discovery of novel thrombin inhibitors. J. Med. Chem. 2006;49:1346–1355. doi: 10.1021/jm050850v. [DOI] [PubMed] [Google Scholar]
- 21.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl Acad. Sci. USA. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]