Abstract
Research results are primarily published in scientific literature and curation efforts cannot keep up with the rapid growth of published literature. The plethora of knowledge remains hidden in large text repositories like MEDLINE. Consequently, life scientists have to spend a great amount of time searching for specific information. The enormous ambiguity among most names of biomedical objects such as genes, chemicals and diseases often produces too large and unspecific search results. We present GeneView, a semantic search engine for biomedical knowledge. GeneView is built upon a comprehensively annotated version of PubMed abstracts and openly available PubMed Central full texts. This semi-structured representation of biomedical texts enables a number of features extending classical search engines. For instance, users may search for entities using unique database identifiers or they may rank documents by the number of specific mentions they contain. Annotation is performed by a multitude of state-of-the-art text-mining tools for recognizing mentions from 10 entity classes and for identifying protein–protein interactions. GeneView currently contains annotations for >194 million entities from 10 classes for ∼21 million citations with 271 000 full text bodies. GeneView can be searched at http://bc3.informatik.hu-berlin.de/.
INTRODUCTION
Scientific literature is the primary medium for communicating novel research results. However, the amount of accumulated texts has reached a point where searching specific information becomes cumbersome. Besides the shear number of available texts, the high ambiguity among most names of biomedical objects such as genes, chemicals and diseases often produces too large and unspecific search results. This is documented by the fact that over one-third of all PubMed queries results in >100 citations (1). Further more, biomedical publications hardly follow naming conventions for entities and remain attached to their authors favorite names (2). Manually assigned keywords such as MeSH terms only partly alleviate the problem, as those tags are notoriously incomplete and represent rather coarse-grained concepts. To address these issues and facilitate entity-specific search over biomedical text repositories, tools are needed that extract semantic knowledge from biomedical literature. In this article, we present GeneView, a web-based application providing access to a comprehensively annotated version of ∼21 million PubMed abstracts and ∼271 000 openly available PubMed Central full texts. It uses a multitude of state-of-the-art text-mining tools optimized for recognizing mentions from 10 different entity classes and for automatically identifying protein–protein interactions (PPI). Among other entities, GeneView currently contains 32.8 million genes, 73.3 million chemicals, 914 000 Single Nucleotide Polymorphisms SNP and 3.9 million PPIs. Besides its broad coverage of entities, GeneView also offers a number of unique features for searching articles. For instance, users may rank query results based on the content of articles with respect to a personalized gene list or by the number of genes or SNPs they contain.
Related work
Several web-based tools exist that support the retrieval of biomedical information using text mining. We discuss those tools that are most similar to GeneView and refer to references (3,4) for excellent reviews of this field. iHop (5) provides access to a subset of PubMed sentences containing at least two proteins in conjunction with interaction specific keywords. Entities other than proteins are not considered. AliBaba (6) aggregates extracted knowledge across all results of a PubMed query and visualizes them as a graph, while GeneView focuses on the individual documents. EbiMed (7) retrieves co-occurring entities for a specific query and ranks them by frequency. Like AliBaba and unlike GeneView, it thus provides aggregated results; furthermore, GeneView uses a sophisticated machine learning technique to detect relationships instead of co-occurrences. UKPMC (8) extends the functionality of PubMed Central by using Whatizit (9) to recognize and highlight entities in abstracts. The system does neither highlight entities in full texts nor does it provide functionality to search with database identifiers instead of (possibly ambiguous) entity names. Finally, GoPubMed (10) recognizes genes, gene ontology and MeSH terms and presents search results using the structure behind these vocabularies. In contrast, GeneView recognizes a broader set of entity types but not gene ontology terms, provides search facilities using unique database identifiers and also finds relationships between proteins in texts. Note that GeneView, in contrast to all systems (except for UKPMC), includes the complete open PMC full text corpus on top of all Medline abstracts. GeneView offers all annotations as downloads to support the development of new applications by freeing developers of data analysis algorithms from the necessity to deal with a multitude of text-mining packages.
SYSTEM DESCRIPTION
GeneView contains all articles from PubMed and the PubMed Central open access subset. To semantically enrich these articles and provide convenient user access, GeneView uses several inter-operating components: (i) named entity recognition and PPI extraction modules; (ii) an inverted index for efficient searching; (iii) a customizable ranking algorithm taking the extracted entity-centric information into account and (iv) a web front end for querying and visualization.
Named entity recognition
For the recognition of named entities, we use a multitude of state-of-the-art tools.
Genes
For gene name recognition and normalization, we use GNAT (11). GNAT is based on custom dictionaries and conditional random fields (CRFs) and normalizes gene mentions to Entrez Gene IDs. The system was ranked among the first in several critical evaluations (12,13) and achieves, according to these assessments, a precision of 82% and a recall of 82% for abstracts and precision/recall values of 54/47% for full-text articles. However, these evaluations were performed on the document level, thus the performance of GNAT at the mention level, as shown in GeneView, might be different. Using local context profiles and heuristics, GNAT tries to find the most probable Entrez Gene ID for a recognized gene. In uncertain cases, GNAT associates gene mentions with more than one ID. Because of different context profiles, a gene mention can be annotated in one sentence, but missed in another one. Annotations are therefore propagated to previously missed tokens including mentions of abbreviations and long-forms.
Single nucleotide polymorphisms
Natural and artificial SNPs are detected using an improved version of MutationFinder (14). This version achieves a precision and recall of 97.5% and 80.7%, respectively, on the original test set of 508 abstracts. Subsequently, we normalize SNP mentions to dbSNP identifiers. Our normalization procedure achieves a precision of 93.0% and a recall of 51.0% on a corpus of 296 documents (15). Mentions of dbSNP identifiers are recognized using regular expressions, achieving a precision of 98.2% on a set of 100 randomly selected documents.
Species
Species are identified and normalized to the NCBI taxonomy using LINNAEUS, which achieves a precision of 97% and recall of 94% on a test corpus of 100 full text documents (16).
Chemicals
We recognize chemical compounds using ChemSpot (17), a hybrid approach using CRF for the detection of IUPAC-like chemical names and a custom dictionary for other chemicals, including trivial names, abbreviations and molecular formulas. ChemSpot achieves a precision of 68% and a recall of 69.5% on the SCAI corpus (18).
Histone modifications
Histone modifications are recognized using a set of 134 regular expressions and normalized to the Brno histone modification nomenclature. This approach achieves a precision of 94.4% and a recall of 88.7% on an evaluation corpus of 1 000 documents (19).
Other named entities
Finally, mentions of cell-types, diseases, drugs, enzymes and tissues are extracted using dictionaries provided by AliBaba (6). Mostly due to the lack of appropriate corpora, it is not possible to provide sensible quality metrics for these classes of entities.
Considering the small size of available corpora, mentioned evaluation values have to be considered as rough estimates. Overlapping annotations, e.g. amino acids inside of mutation mentions, are not disambiguated. Instead, link outs are provided for all these instances. This also holds for recognized gene names where GNAT was unable to distinguish between several Entrez Gene IDs.
GeneView uses a machine learning approach based on support vector machines for relationship extraction between recognized proteins (20). The final model is trained on the ensemble of five corpora annotated with PPIs (21). The method achieved very good results in a comprehensive evaluation of nine machine learning approaches for PPI extraction (22). Depending on the evaluation corpus, F1 scores from 54.5% to 74.5% are observed.
The web interface currently presents results for genes, SNPs, chemicals, histone modifications, drug names and PPIs. These and additional entity types are provided as separate download. The document repository of GeneView is updated on a regular basis of 3 months and annotations are renewed when major releases of the NER tools are published.
Indexing
All PubMed abstracts and freely available PMC full texts are downloaded as XML files, parsed and imported into Lucene. Lucene serves GeneView as text storage, query processor and ranking engine. For each article, we parse additional information such as author names, affiliations, journal, MeSH terms and date of publication. Sections of full-text articles (e.g. title, methods, result and figure/table caption) are identified by a dictionary containing commonly used section names. This enables users to restrict queries to certain parts of an article. Named entity recognition and relation extraction are applied on all texts and stored in a relational database to allow for structured retrieval and aggregation.
Ranking
The range of indexed information allows to provide several advanced methods for ranking search results. These include rankings by relevance regarding the user query, date of publication or the numbers of entities of a certain type (genes and SNPs) the articles contain. Ranking by relevance is based on term frequencies in the articles, respective selectivity/importance of the given query terms and the average section length. Thus, short sections such as ‘title’ or ‘abstract’ are considered to be more important than longer sections like ‘methods’. We use section-specific boosting for gene queries, with the highest relevance score assigned to gene mentions in ‘title’, ‘abstract’, or ‘result’ and the lowest to those mentioned in ‘methods’ or ‘introduction’. Query results can also be ranked by a user-defined gene list (see third use case below).
Interface
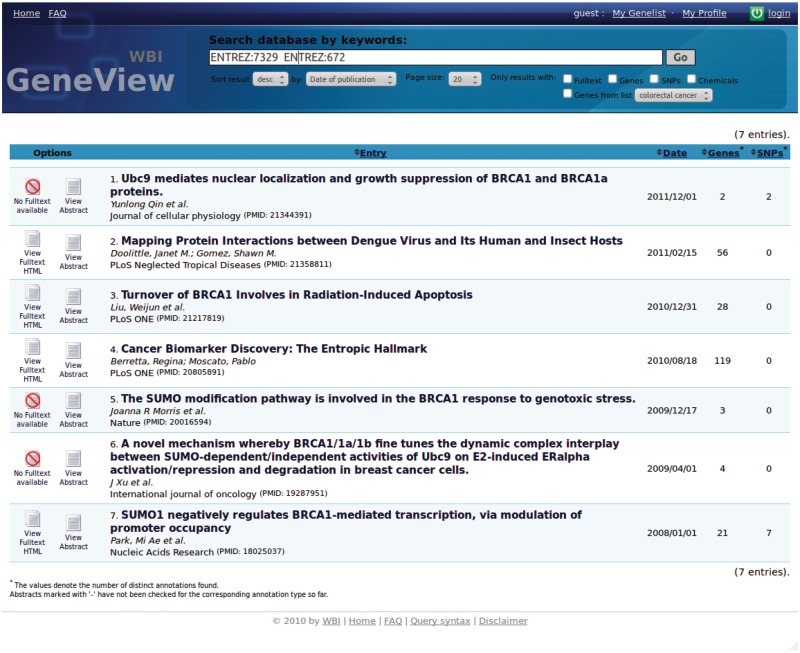
GeneView is accessible through an intuitive web-based interface. The central point of access is a search form which supports several types of queries. These queries can take full advantage of the collected underlying data and advanced ranking options. Performing a search returns a list of matching articles. By default, these articles are ranked by publication date. Previously described ranking options, e.g. ranking by relevance or entity count, are available for user selection. In addition, a user can reduce the number of retrieved articles by filtering for mentions of genes, mutations or chemical compounds. The result of a query for two genes (UBE2I and BRCA1) is displayed in Figure 1.
Figure 1.
Query result for articles mentioning genes UBE2I (GeneID 7329) and BRCA1 (GeneID 672).
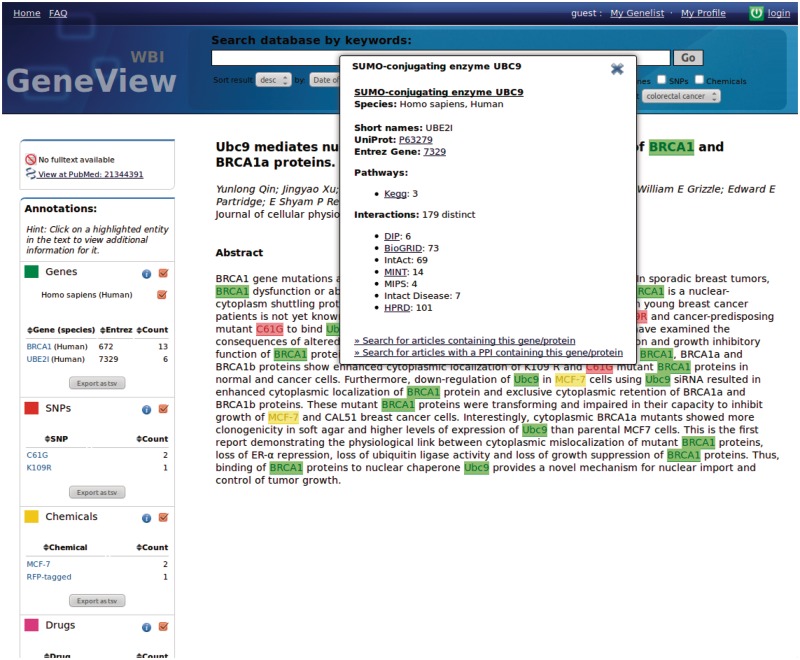
The visualization of an article selected from the search results is shown in Figure 2. The view is separated into two panels. The main, right-hand panel shows the article, including authors and journal. A mouse click triggers a query for publications from the selected author or journal. Entity markup is directly provided in the text and additional entity-specific information is displayed in a pop-up on mouse click. This encompasses link outs to entity-specific databases (e.g. UniProt, DrugBank or IntAct) or additional information such as the official gene name. The markup also provides one-click functionality to search for articles discussing the selected entity. For gene names, this includes the possibility to search specifically for those articles where the binary classifier detected a PPI for the selected entity. The respective sentence is provided on user request.
Figure 2.
Visualization of a selected article (PMID 21344391). Additional information such as full gene name and links to external databases can be provided for a selected entity.
The left-hand panel shows an overview of all entities found in the article sorted by overall frequency. Per default, all entities are highlighted, but users may also restrict coloring to certain entity types. For gene names, the markup can also be restricted by species. Tokens annotated by more than one tool are highlighted using an intermediate color and a mouse click provides all available information.
RESULTS AND DISCUSSION
GeneView currently contains 20 962 294 abstracts together with 271 808 full-text bodies. All articles are automatically annotated by 10 named entity recognition tools. As of January 2012, the repository contains >194 million entities (for a thorough overview, see Table 1). Out of 15 197 637 co-occurring protein mentions, 3 921 267 (25.8%) are classified as PPI. The availability of full texts directly in GeneView provides an additional source of information. For instance, >21 400 of ∼172 000 articles contain SNPs in the full text only. Similarly, the overall number of SNPs reduces from 914 543 to 523 398 (57.2%) after ignoring full-text mentions. These numbers emphasize the much better coverage of full-text articles.
Table 1.
Overview of all entities in the GeneView repository
| Entity type | Articles | Entities | Unique | Normalized |
|---|---|---|---|---|
| Chemical | 9 851 347 | 73 354 240 | 59 232 | ChemIDPlus |
| Species | 8 815 334 | 40 992 161 | 110 880 | NCBI Taxonomy |
| Drug | 6 023 081 | 44 595 216 | 3 052 | DrugBank/PharmGKB |
| Gene | 2 855 898 | 32 861 120 | 81 229 | Entrez Gene |
| Enzyme | 561 152 | 825 889 | 2 519 | Kegg |
| Disease | 272 240 | 679 364 | 9 681 | MeSH |
| SNP | 171 597 | 914 543 | 18 942 | dbSNP |
| Cell-type | 36 851 | 82 285 | 585 | MeSH |
| Tissue | 8 164 | 9 488 | 132 | MeSH |
| Histone Mod. | 5 938 | 62 370 | 316 | Brno nomenclature |
Articles: number of citations with at least one entity found; entities: total number of recognized mentions; unique: number of distinct entities; normalized: identifier mentions are normalized to.
Functionality and usage
The functionality of GeneView is described by the following four use cases.
Entity-specific search
Ambiguity of entity names is a well-studied problem for several entity types. For example, the acronym ‘PAP’ refers to more than eight different human genes but also to concepts such as ‘pulmonary artery pressure’. Conversely, a single entity may be referred to by multiple different names. By using named entity recognition and normalization, GeneView greatly reduces the impact of such problems and facilitates finding of relevant articles. GeneView also allows to query articles using unique identifiers such as Entrez Gene for genes, dbSNP for SNPs, ChemIDPlus for chemicals, Brno abbreviations for histone modifications and DrugBank or PharmGKB for drugs.
The advantage of GeneView's entity normalization capabilities becomes apparent when searching for articles describing, say, a specific SNP. Assume a researcher searches for the schizophrenia-associated SNP Val158Met located on the gene COMT. A direct search for this name leads to 448 hits in PubMed but ignores the high number of lexical variations (e.g. V158M, Val158→Met, Val(158)Met, and Val158/Met). Lexical variations are recognized and unified in GeneView, yielding 575 articles in return for a search for V158M. Still, such a search neglects the high number of possibilities to describe SNP (e.g. Val108Leu, 472G>A, and 322G>C) and the fact that V158M is known to exist for almost 20 different human genes. Only a search using the unique dbSNP identifier rs4680 deals with these problems: a search for rs4680 returns 672 articles. In comparison, dbSNP itself covers references to only 165 PubMed articles for this SNP.
The result of such a search in GeneView can be combined with all filtering and ranking options previously described. Also, entity-specific queries can be combined with any kind of keyword-based or another entity-specific search. This allows the user to form complex queries, for example, to search for a specific SNP co-mentioned with a gene. Such a search can be useful when the SNP of interest is not contained in dbSNP or the identifier is unknown. For example, GeneView returns 531 articles when searching for the SNP V158M in conjunction with the gene COMT. A basic overview of co-occurring entities is shown in Table 2.
Table 2.
Number of co-occurring concepts contained in GeneView
| Entity 1 | Entity 2 | Co-occurrence |
|---|---|---|
| Gene | Chemical | 48 278 038 |
| Gene | Drug | 20 099 049 |
| Gene | SNP | 1 203 334 |
| Gene | Histone modification | 162 108 |
| SNP | Chemical | 3 270 485 |
| SNP | Drug | 1 214 063 |
| SNP | Histone modification | 5 267 |
Multiple mentions of the same entity are only counted once.
Advanced keyword-based search
Revisiting our initial example, a researcher might be interested in an overview of SNPs associated with schizophrenia. A search for ‘schizophrenia’ leads to >63 000 articles. Using GeneView to restrict the search to articles mentioning at least one SNP reduces this result to ∼1 700 articles. Ranking by the number of SNPs allows a user to find those articles discussing a high number of different SNPs in conjunction with schizophrenia. Similar queries can also be performed for other entity types of interest. In addition to regular term and phrase searches known from other search engines, the integration of context-related information enables section-specific searching. For instance, queries such as ‘find all articles where a “caption” contains the gene EGFR in conjunction with the phrase western blot’ are possible.
Gene list
Researchers often have a dedicated list of genes they are interested in. GeneView provides users with the possibility to maintain a personalized list of relevant genes and allows to restrict query results to articles containing at least one of these genes of interest. Matching articles can be scored and ranked according to these genes of interest. The scoring modifies the default ranking strategy by incorporating the section where genes appear.
We demonstrate the utility of this feature by the following example: GeneView returns 62 798 articles matching the query term ‘schizophrenia’. We added all 26 genes from OMIM associated with the disease schizophrenia into our personal gene list. Using this user-defined gene list as a filter allows to reduce the query result to 1 269 articles containing the keyword and at least one gene of interest. Changing the ranking to ‘relevance’ pulls articles discussing the impact of several of these genes on schizophrenia to the top of the result listing.
Biocuration
An ongoing challenge is to complete the functional annotation of genes. Baumgartner et al. (23) estimate that the annotation process for all human genes with at least one ‘Gene Reference Into Function’ tag will not be finished before 2020, unless technological advancements can improve the annotation process considerably. Augmentation of articles with automatically derived annotations may substantially decrease the time needed to read and, if necessary, to curate an article (24). By the multitude of annotated entities, GeneView is perfectly suited to support manual curation of large biomedical databases by selectively searching and augmenting biomedical articles. GeneView can also be used by researchers to find PPIs to augment existing PPI networks.
Download
All annotations are also available as structured downloads. This enables large-scale analysis such as the retrieval of co-occurring terms, the integration of text-mining results in other tools or the possibility to analyze the long-term development of biomedical concepts. This is exemplified for the genes BRCA1 and BRCA2 in conjunction with at least one SNP in Figure 3.
Figure 3.
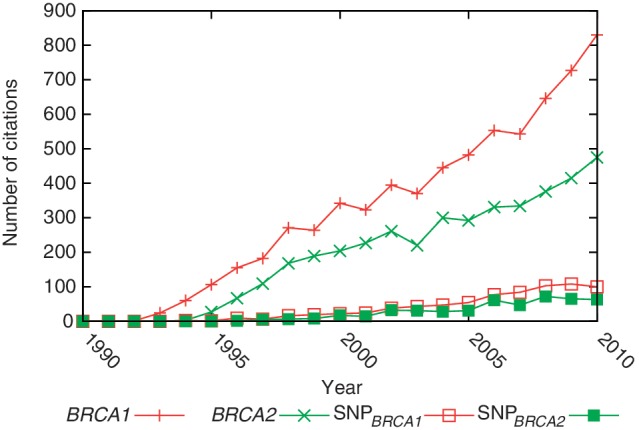
Number of citations where the genes BRCA1 or BRCA2 occur in for the last 20 years. Similarly, we show the progression of articles with SNPs co-mentioned with BRCA1 or BRCA2.
CONCLUSION
Keeping pace with latest research results is becoming more and more difficult for biomedical researchers. We introduce GeneView, a fast and powerful tool for navigating the biomedical literature. The most important features of GeneView include the possibility to search for articles describing a specific biological entity, flexible ranking of result according to the users need using optimized ranking algorithms and an intuitive visualization of semantically annotated texts. Queries are conducted on abstracts and full texts simultaneously. All annotated entities are interactive: a mouse click provides additional information such as links to external reference databases or pathway and interaction information and enables the search for additional articles on this entity directly in GeneView.
GeneView currently provides the most comprehensive semantic search engine for the Life Sciences. GeneView can considerably reduce the necessary effort for searching, reading, understanding and annotating biomedical articles.
FUNDING
German Federal Ministry of Education and Research [0315417B] and German Research Foundation [GRK 1651/1]. Funding for open access charge: German Research Council.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank the reviewers for their careful reading and helpful comments.
REFERENCES
- 1.Dogan RI, Murray GC, Névéol A, Lu Z. Understanding PubMed user search behavior through log analysis. Database. 2009;2009 doi: 10.1093/database/bap018. bap018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tamames J, Valencia A. The success (or not) of HUGO nomenclature. Genome Biol. 2006;7:402. doi: 10.1186/gb-2006-7-5-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lu Z. PubMed and beyond: a survey of web tools for searching biomedical literature. Database. 2011;2011 doi: 10.1093/database/baq036. baq036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rodriguez-Esteban R. Biomedical text mining and its applications. PLoS Comput Biol. 2009;5:e1000597. doi: 10.1371/journal.pcbi.1000597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fernández JM, Hoffmann R, Valencia A. iHOP web services. Nucleic Acids Res. 2007;35:W21–W26. doi: 10.1093/nar/gkm298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Plake C, Schiemann T, Pankalla M, Hakenberg J, Leser U. AliBaba: PubMed as a graph. Bioinformatics. 2006;22:2444–2445. doi: 10.1093/bioinformatics/btl408. [DOI] [PubMed] [Google Scholar]
- 7.Rebholz-Schuhmann D, Kirsch H, Arregui M, Gaudan S, Riethoven M, Stoehr P. EBIMed–text crunching to gather facts for proteins from Medline. Bioinformatics. 2007;23:e237–e244. doi: 10.1093/bioinformatics/btl302. [DOI] [PubMed] [Google Scholar]
- 8.McEntyre JR, Ananiadou S, Andrews S, Black WJ, Boulderstone R, Buttery P, Chaplin D, Chevuru S, Cobley N, Coleman L-A, et al. UKPMC: a full text article resource for the life sciences. Nucleic Acids Res. 2011;39:D58–D65. doi: 10.1093/nar/gkq1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rebholz-Schuhmann D, Arregui M, Gaudan S, Kirsch H, Jimeno A. Text processing through Web services: calling Whatizit. Bioinformatics. 2008;24:296–298. doi: 10.1093/bioinformatics/btm557. [DOI] [PubMed] [Google Scholar]
- 10.Doms A, Schroeder M. GoPubMed: exploring PubMed with the Gene Ontology. Nucleic Acids Res. 2005;33:W783–W786. doi: 10.1093/nar/gki470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hakenberg J, Gerner M, Haeussler M, Solt I, Plake C, Schroeder M, Gonzalez G, Nenadic G, Bergman CM. The GNAT library for local and remote gene mention normalization. Bioinformatics. 2011;27:2769–2771. doi: 10.1093/bioinformatics/btr455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morgan AA, Lu Z, Wang X, Cohen AM, Fluck J, Ruch P, Divoli A, Fundel K, Leaman R, Hakenberg J, et al. Overview of BioCreative II gene normalization. Genome Biol. 2008;9:S3. doi: 10.1186/gb-2008-9-s2-s3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu Z, Kao H-Y, Wei C-H, Huang M, Liu J, Kuo C-J, Hsu C-N, Tsai RT-H, Dai H-J, Okazaki N, et al. The gene normalization task in BioCreative III. BMC Bioinformatics. 2011;12(Suppl 8):S2. doi: 10.1186/1471-2105-12-S8-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Caporaso JG, Baumgartner WA, Randolph DA, Cohen KB, Hunter L. MutationFinder: a high-performance system for extracting point mutation mentions from text. Bioinformatics. 2007;23:1862–1865. doi: 10.1093/bioinformatics/btm235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thomas P, Klinger R, Furlong L, Hofmann-Apitius M, Friedrich C. Challenges in the association of human single nucleotide polymorphism mentions with unique database identifiers. BMC Bioinformatics. 2011;12(Suppl 4):S4. doi: 10.1186/1471-2105-12-S4-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gerner M, Nenadic G, Bergman CM. LINNAEUS: a species name identification system for biomedical literature. BMC Bioinformatics. 2010;11:85. doi: 10.1186/1471-2105-11-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rocktäschel T, Weidlich M, Leser U. ChemSpot: a hybrid system for named entity recognition of chemicals. Bioinformatics. 2012;28:1633–1640. doi: 10.1093/bioinformatics/bts183. [DOI] [PubMed] [Google Scholar]
- 18.Kolářik C, Klinger R, Friedrich CM, Hofmann-Apitius M, Fluck J. Workshop on Building and evaluating resources for biomedical text mining. Marrakech, Morocco: 2008. Chemical names: terminological resources and corpora annotation; pp. 51–58. [Google Scholar]
- 19.Kolářik C, Klinger R, Hofmann-Apitius M. Identification of histone modifications in biomedical text for supporting epigenomic research. BMC Bioinformatics. 2009;10(Suppl 1):S28. doi: 10.1186/1471-2105-10-S1-S28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Giuliano C, Lavelli A, Romano L. Proceedings of the 11th Conference of the European Chapter of the Association for Computational Linguistics (EACL 2006) Trento, Italy: 2006. Exploiting shallow linguistic information for relation extraction from biomedical literature; pp. 401–408. [Google Scholar]
- 21.Pyysalo S, Airola A, Heimonen J, Björne J, Ginter F, Salakoski T. Comparative analysis of five protein-protein interaction corpora. BMC Bioinformatics. 2008;9(Suppl 3):S6. doi: 10.1186/1471-2105-9-S3-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tikk D, Thomas P, Palaga P, Hakenberg J, Leser U. A comprehensive benchmark of kernel methods to extract protein protein interactions from literature. PLoS Comput. Biol. 2010;6:e1000837. doi: 10.1371/journal.pcbi.1000837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baumgartner WA, Cohen KB, Fox LM, Acquaah-Mensah G, Hunter L. Manual curation is not sufficient for annotation of genomic databases. Bioinformatics. 2007;23:i41–i48. doi: 10.1093/bioinformatics/btm229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arighi C, Roberts P, Agarwal S, Bhattacharya S, Cesareni G, Chatr-aryamontri A, Clematide S, Gaudet P, Giglio M, Harrow I, et al. BioCreative III interactive task: an overview. BMC Bioinformatics. 2011;12(Suppl 8):S4. doi: 10.1186/1471-2105-12-S8-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]