Abstract
DR_bind is a web server that automatically predicts DNA-binding residues, given the respective protein structure based on (i) electrostatics, (ii) evolution and (iii) geometry. In contrast to machine-learning methods, DR_bind does not require a training data set or any parameters. It predicts DNA-binding residues by detecting a cluster of conserved, solvent-accessible residues that are electrostatically stabilized upon mutation to Asp−/Glu−. The server requires as input the DNA-binding protein structure in PDB format and outputs a downloadable text file of the predicted DNA-binding residues, a 3D visualization of the predicted residues highlighted in the given protein structure, and a downloadable PyMol script for visualization of the results. Calibration on 83 and 55 non-redundant DNA-bound and DNA-free protein structures yielded a DNA-binding residue prediction accuracy/precision of 90/47% and 88/42%, respectively. Since DR_bind does not require any training using protein–DNA complex structures, it may predict DNA-binding residues in novel structures of DNA-binding proteins resulting from structural genomics projects with no conservation data. The DR_bind server is freely available with no login requirement at http://dnasite.limlab.ibms.sinica.edu.tw.
INTRODUCTION
Interactions between proteins and DNA play essential roles for life. For example, protein–DNA interactions control gene regulation, cell replication and transcription, as well as DNA repair. Furthermore, many of these DNA-binding proteins are involved in human diseases such as neurological disorders, e.g. TDP-43 (1), and cancer; e.g. p53 (2). Consequently, identifying the key amino acid residues involved in DNA recognition is critical for understanding these important biological processes. It also guides which residues to mutate in experimental studies.
Several methods and web servers have been developed to predict DNA-binding residues from the protein 1D sequence or 3D structure. Methods that predict DNA-binding residues using only the protein sequence generally employ machine-learning algorithms such as a neural network (3–5), a Naïve Bayes classifier (6), a support vector machine (7–12), random forest (13,14), or decision trees (C4.5 algorithm) (15). These algorithms usually employ amino acid physicochemical properties, sequence conservation, the local sequence context, solvent accessibility and/or secondary structure. Publicly available web servers that implement sequence-based methods for predicting DNA-binding residues include DBS-PRED (3), DBS-PSSM (5), DNABindR (6), DP-Bind (8), DISIS (9), BindN-rf (14), BindN+ (12), NAPS (15) and MetaDBSite (16). Methods that use the protein structure, if available, generally improve the DNA-binding site prediction, as they replace the predicted solvent accessibility, hydrophobicity and secondary structure in sequence-based methods with observed ones and can additionally employ energies or frequencies, computed from the atomic coordinates, as well as experimental geometrical features. Structure-based methods for predicting DNA-binding residues employ mostly electrostatic potentials in conjunction with other features such as surface/solvent accessibility, the protein surface shape, amino acid conservation, propensity, hydrophobicity and hydrogen-bonding potential and structural motifs (17–22), or high-frequency residue fluctuations (23). Servers that implement structure-based methods for predicting DNA-binding residues include PreDs (24), DISPLAR (25), DBD-Hunter (26) and DNABINDPROT (23).
In our previous work (27), we had developed a structure-based DNA-binding residue prediction method based on (i) electrostatics, (ii) conservation and (iii) geometry with the following rationale: (i) DNA-binding residues contain electropositive atoms, which would be in an unfavorable electrostatic environment in the absence of DNA or water; thus replacing one of these residues with a negatively charged Asp−/Glu− would alleviate the electrostatic repulsion among the electropositive atoms in the gas phase; (ii) DNA-binding residues and residues in the vicinity, which form a cluster of spatially interacting residues, are usually highly conserved within the same family due to their critical functional roles and (iii) DNA-binding residues have been observed to be located on surface patches, as opposed to clefts/cavities for RNA-binding residues and enzyme substrates. In this work, we have implemented our DNA-residue prediction method for public use in a web server, DR_bind (http://dnasite.limlab.ibms.sinica.edu.tw). Whereas our published method for predicting DNA-binding sites had been tested on a non-redundant set of 56 DNA-bound and 23 DNA-free non-homologous protein structures (27), DR_bind was tested herein using an updated non-redundant set of 83 DNA-bound and 55 DNA-free structures (referred to as Data sets I and II, respectively). DR_bind was also tested using a protein–DNA docking benchmark containing 47 unbound–bound structures (28) and 15 non-redundant DNA-bound protein structures with no or insufficient homologous seqeunces to compute conservation scores reliably. In contrast to current DNA-binding residue prediction servers, DR_bind is based on physical principles of binding thermodynamics (29) and does not require training on a set of protein–DNA complexes or any parameters. Hence, DR_bind would be an opportune addition since structures of DNA-binding proteins have been rapidly rising.
METHODS
Data sets used
DR_bind was tested using four data sets: I—83 non-redundant DNA-bound protein structures, II—55 non-redundant DNA-free protein structures, III—47 bound–unbound structures from the protein–DNA benchmark version 1.2 (28) and IV—15 non-redundant DNA-bound protein structures with no, or insufficient homologs to compute conservation profiles reliably. To create Data set I, all available X-ray structures of DNA-bound proteins solved to ≤3-Å resolution were obtained from the current Protein Data Bank (PDB) (30). These protein chains were grouped according to their Class, Architecture, Topology and Homologous superfamily (CATH) codes (31). For each group of protein structures with the same CATH code, the structure with the best resolution was selected as the representative one. If any of these representative proteins share >30% sequence identity, the protein with the longer sequence was kept, while the others were discarded. This yielded 83 DNA-bound proteins that are sequentially and structurally non-homologous with conservation data (Supplementary Table S1), whereas the remaining 12 proteins had no conservation profiles from the ConSurf-DB database (http://consurfdb.tau.ac.il/) (32).
Data set II was derived from Data set I by searching each of the 83 DNA-bound proteins with conservation data for highly homologous proteins (sharing ≥90% sequence identity) with DNA-free structure(s) using the SAS tool (http://www.ebi.ac.uk/thornton-srv/databases/sas/); if multiple DNA-free structures were found, the structure that showed the largest root-mean-square deviation (RMSD) from the DNA-bound structure using the SSAP program (33) was chosen as the representative one. This yielded 55 bound–unbound structures with a wide range of RMSDs (0.3–33 Å). The PDB entries of the DNA-bound and free protein structures, the sequence identity between the DNA-bound and the respective free proteins computed using global alignment with ClustalW1.83 (34) and their RMSD values are given in Supplementary Table S1.
Data set III is a protein–DNA docking benchmark containing 47 bound–unbound structures, of which 13 were classified as ‘easy’, 22 as ‘intermediate’ and 12 as ‘difficult’ cases for docking depending on the interface RMSD values between the DNA-bound and corresponding free structures. ‘Easy’, ‘intermediate’ and ‘difficult’ structures were defined by interface RMSD values ranging from 0 to 2 Å, 2 to 5 Å, >5 Å, respectively. Data set III differs from Data set II in that it includes: (i) protein structures deposited in the September 2007 RCSB PDB; (ii) structurally homologous proteins with the same CATH code; (iii) free NMR structures; and (iv) 15 structures without conservation data from ConSurf-DB.
To create Data set IV, the 12 proteins excluded from Data set I and the 15 proteins from the benchmark set, which lack conservation profiles from ConSurf-DB, were grouped according to their CATH codes. For each group of protein structures with the same CATH code, the best resolution structure was selected as the representative one. This yielded 15 non-redundant proteins sharing <30% pairwise sequence identity (Supplementary Table S2).
Definitions
A residue was considered to bind DNA if it contains one or more non-hydrogen atom within van der Waals contact or hydrogen-bonding distance to the non-hydrogen atom of its binding partner directly or indirectly via a bridging water molecule. HBPLUS (35) was used to compute all possible hydrogen bonds and van der Waals contacts, which are defined by a donor atom to an acceptor atom distance ≤3.5 and ≤4.0 Å, respectively. An amino acid X is considered accessible for interacting with DNA if the percent ratio of its side chain solvent-accessible surface area in the protein to that in the tripeptide, –Gly–X–Gly–, is >5% (17,36). MOLMOL (37) was used to compute the relative solvent-accessible surface area of each amino acid from the protein structure using a solvent probe radius of 1.4 Å.
Geometry
Since DNA-binding sites are found on a protein surface, surface patches were generated by defining the Cα atom of each residue as an origin of a patch and including all residues whose Cα atoms were within 10 Å of the origin in the patch. Non-identical patches with more than five solvent-accessible residues were used in computing the average electrostatic energy change and conservation (see below).
Electrostatics
Given a l-residue DNA-binding protein structure, all Asp/Glu residues were deprotonated, while Arg/Lys residues were protonated; His residues were protonated or deprotonated depending on the availability of hydrogen bond acceptors in the structure. Next, l mutant structures were generated by replacing Ala, Asn, Asp, Cys, Gly, Ser, Thr or Val in the wild-type structure to Asp− and the other residues to Glu−. The side chain replacements were carried out using SCWRL (38), followed by energy minimization with heavy constraints on all heavy atoms using AMBER (39) to relieve any bad contacts. Based on the wild-type/mutant structures, the gas-phase (ε = 1) electrostatic energy of the wild-type (Eelecwt) or mutant (Eelecmut) protein in the ‘folded’ state relative to that in an ‘extended reference’ state (E′elecwt or E′elecmut) was computed using AMBER (39) with the all-hydrogen-atom AMBER force field (40). In this extended reference state, the residues do not interact with one another; hence, the electrostatic energy difference between the wild-type (E′elecwt) or mutant (E′elecmut) ‘unfolded’ protein is equal to the difference between the electrostatic energies of the native residue at position i (E′eleci) and the corresponding mutant Asp−/Glu− (E′elecD/E). The change in the gas-phase electrostatic energy ΔΔEeleci, upon mutation of residue i to Asp−/Glu− is given by:
| (1) |
The average electrostatic energy change <ΔΔEelec>i of the Naai residues comprising surface patch i was computed from:
| (2) |
where the summation in Equation (2) is over all residues in patch i.
Conservation
For a given DNA-binding protein, the conservation score Ci of residue i was obtained from the ConSurf-DB database (32) or ConSurf server (41–43). The Ci score is an integer number, ranging from 1 (for a rapidly evolving, highly variable residue) to 9 (for a slowly evolving, conserved residue). The average conservation <C>i of the Naai residues comprising surface patch i was computed from:
| (3) |
DNA-binding residue prediction
To determine the DNA-binding residues in a given protein, the distinct patches were ranked according to the <ΔΔEelec>i values so that the top-ranked cluster had the most favorable (most negative) <ΔΔEelec>i, whereas the bottom-ranked cluster had the least favorable <ΔΔEelec>i . Among the top 10% <ΔΔEelec>i-ranked surface patches, the three patches with the largest <C>i values were selected and the constituent solvent-accessible residues were predicted to bind DNA.
Performance measures
To evaluate the performance of DR_bind, the numbers of correctly predicted binding residues (TP) and non-binding residues (TN), as well as the numbers of incorrectly predicted binding residues (FP) and non-binding residues (FN) were computed and used to determine:
| (4) |
| (5) |
| (6) |
| (7) |
 |
(8) |
DR_bind web server
Input
On the DR_bind web page http://dnasite.limlab.ibms.sinica.edu.tw/, users are given two options: For option A, users upload their own file in PDB format and the evolutionary data for their protein in ConSurf format or ask DR_bind to retrieve the evolutionary data from ConSurf. For option B, users enter the PDB code and chain identifier; if the conservation profile for the submitted protein structure has not been pre-calculated in the ConSurf-DB database (32), DR_bind will attempt to generate the ConSurf data automatically from the ConSurf server (41–43). If no ConSurf data can be generated, DR_bind will continue to predict DNA-binding residues based only on the protein 3D structure and inform the user of the missing ConSurf data on the Results page. For multiple submissions, we have provided a simple form that allows for nine PDB codes with chain identifiers to be defined. After users click on the ‘submit’ button, the input data is checked for consistency: Residues in the PDB file that do not correspond to the standard 20 amino acid are removed, as well as multiple alternative residue positions. If the input data pass these tests, then the prediction process is started and the user is taken to a web page where the results for the job(s) and their status on the DR-bind server can be monitored.
Output
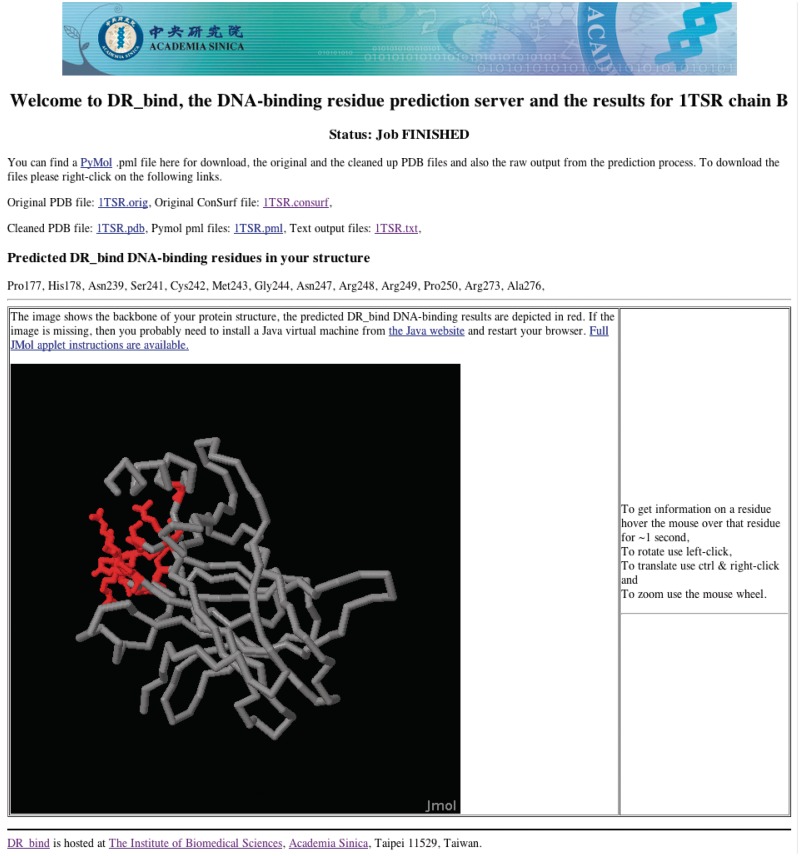
When the DR_bind server has finished the prediction, the results page is updated with the predicted binding residues. If the user had provided an e-mail address, the web server will send an e-mail to let the user know that the prediction has been completed with a link to the results web page. Users can then access the results page to see the generated prediction. As shown in Figure 1, the results page is split into three sections: the first section has links to downloadable files of (i) the original PDB and ConSurf files, (ii) the ‘cleaned’ PDB file used by DR_bind, (iii) a PyMOL script for highlighting the predicted DNA-binding residues and (iv) a text file of these residues. The second section lists the predicted DNA-binding residues. The third section is an interactive embedded 3D representation of the protein with the entire backbone in ribbon format with the predicted interaction residues depicted in stick format in red. This 3D representation is created using Jmol (http://www.jmol.org/) and can be rotated and zoomed in/out on the results page itself.
Figure 1.
An example of the Results page from DR_bind.
DR_bind currently runs on an Apple Mac Mini quad-core i7 server and the time taken to yield a prediction depends on the number of residues in the PDB chain. A prediction takes ∼5 min for 50 residues, ∼1.5 h for 200 residues, ∼4.5 h for 350 residues and ∼10 h for 450 residues. To handle simultaneous requests, the Torque batch processing software is used to queue jobs. Help pages with instructions on how to use the server are available at http://dnasite.limlab.ibms.sinica.edu.tw/examples/help.html.
RESULTS AND DISCUSSION
Performance and limitations of DR_bind
In our previous works (27), we presented a method for predicting DNA-binding sites based on electrostatics, conservation and geometry given the respective protein structure and tested it on a set of 56 structurally non-homologous proteins with DNA-bound structures, as well as a smaller subset of 23 proteins with both DNA-bound and free structures. Based on the DNA-free and DNA-bound protein structures, 83 and 86% of the DNA-binding proteins have statistically significant DNA-binding sites, respectively. Thus, the method was found not to be very sensitive to protein conformational changes upon DNA binding (27,44). However, like all structure-based prediction methods, it cannot predict binding residues in regions that are disordered in the free protein structure. Another limitation of the method is that the predicted residues may be involved in binding non-DNA ligands such as RNA, protein, small molecules or metal ions rather than DNA (27,44).
In this work, we have implemented our DNA-binding residue prediction method as a free web server called DR_bind, which requires as input, the protein 3D structure and yields as output, experimentally testable residues that are predicted to bind DNA. As more DNA-binding protein structures have been solved since validation of our method (27), and some of these may correspond to novel folds, DR_bind was further tested using our updated set of 83 DNA-bound and 55 bound–unbound non-homologous protein structures, as well as the protein–DNA benchmark version 1.2 containing 47 bound–unbound structures (28). DR_bind yielded 47% precision, 35% sensitivity, 96% specificity, 90% accuracy and 35% mcc in predicting DNA-binding residues using our bound data set, and slightly lower precision (43%) and mcc (33%) values using our free data set (Table 1), even though the RMSD of the DNA-free structure from the respective DNA-bound structure may be as large as 33 Å (Supplementary Table S1). Similar trends were found for the benchmark data set: DR_bind yielded 56% precision, 40% sensitivity, 95% specificity, 87% accuracy and 40% mcc using the DNA-bound structures and lower precision (49%) and mcc (35%) values using the corresponding free structures (Table 1). The sensitivity values are low, as DR_bind predicts the most likely DNA-binding residues, rather than all DNA-binding residues at the protein–DNA interface.
Table 1.
Comparison of the performance measures of DR_Bind using our nonredundant data set of 83 DNA-bound and 55 DNA-free protein structures and the protein–DNA benchmark version 1.2 containing 47 DNA-bound and free protein structures
| Data set | I (bound) | II (free) | III (bound) | III (free) |
|---|---|---|---|---|
| No. of structures | 83 | 55 | 47 | 47 |
| TP | 728 | 419 | 468 | 417 |
| FP | 831 | 566 | 371 | 429 |
| TN | 18 128 | 11 596 | 6486 | 6435 |
| FN | 1,362 | 792 | 702 | 693 |
| Precision | 0.47 | 0.43 | 0.56 | 0.49 |
| Sensitivity | 0.35 | 0.35 | 0.40 | 0.38 |
| Specificity | 0.96 | 0.95 | 0.95 | 0.94 |
| Accuracy | 0.90 | 0.90 | 0.87 | 0.86 |
| mcc | 0.35 | 0.33 | 0.40 | 0.35 |
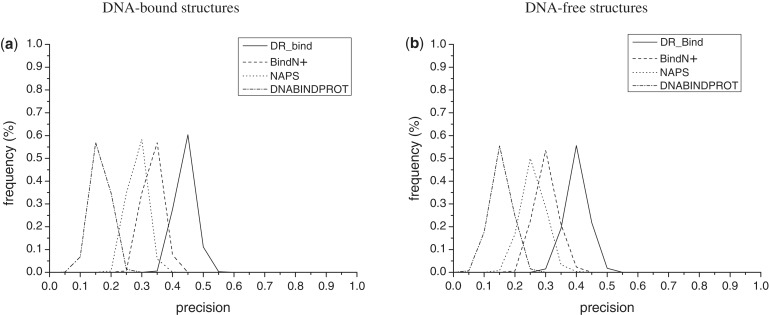
To assess the reliability of the performance values in Table 1, we randomly chose 40 of the 83 DNA-bound structures and 25 of the 55 DNA-free protein structures and computed the various performance measures; this procedure was repeated 1000 times in order to obtain the distribution of each performance measure. Figure 2a and b illustrates the percent frequency of the DR_bind’s precision values (solid lines) for the bound and free data sets, respectively. The lower limits of precision, sensitivity, specificity, accuracy and mcc in predicting DNA-binding residues using DR_bind for the bound/free data sets are 0.38/0.31, 0.29/0.26, 0.94/0.93, 0.87/0.86 and 0.29/0.24, whereas the corresponding upper limits are 0.56/0.55, 0.44/0.49, 0.97/0.97, 0.91/0.92 and 0.43/0.44. Notably, these limits encompass the precision, sensitivity, specificity, accuracy and mcc values obtained using the 47 bound–unbound structures from the benchmark data set.
Figure 2.
The percent frequency of a precision value derived from 1000 random choices of (a) 40 DNA-bound structures from Data set I and (b) 25 DNA-free protein structures from Dataset II. The solid, dashed, dotted and dashed–dotted curves correspond to precision values obtained using DR_bind, BindN+, NAPS and DNABINDPROT, respectively.
Comparisons with other servers that predict DNA-binding residues
Using our bound and free data sets, the performance of DR_bind was compared with that of three recent web servers, BINDN+ (http://bioinfo.ggc.org/bindn+/), NAPS (http://proteomics.bioengr.uic.edu/NAPS) and DNABINDPROT (http://www.prc.boun.edu.tr/appserv/prc/dnabindprot/). BINDN+ (12) uses support vector machines with three biochemical features (hydrophobicity, side chain pKa and mass of an amino acid residue) incorporating evolutionary information and position-specific scoring matrix (PSSM). Instead of support vector machines, NAPS (15) employs ensemble classifiers based on C4.5, bootstrap aggregation and a cost-sensitive learning algorithm with residue charge and PSSM. Whereas BINDN+ and NAPS are sequence-based methods, DNABINDPROT (23) is a structure-based method that identifies high-frequency fluctuating conserved residues and ranks them according to their DNA-binding propensity. These web servers were chosen for comparison with DR_bind because they had been tested using published data sets and had been shown to outperform previous methods/web servers: Using the PDNA-62 data set, the average of sensitivity and specificity obtained by BINDN+ (78.3%) and NAPS (78.5%) were similar (12,15) and higher than that obtained by DP-Bind (76.5%) or DBS-PSSM (67.1%). Using a set of 36 DNA-binding proteins with both free and DNA-bound structures and conservation scores, the precision obtained by DNABINDPROT using a fast threshold of 0.1, conservation threshold of 5, and neighboring two residues (45.3%) was higher than that obtained by DBD-HUNTER (44.5%), DISPLAR (40%) and DP-Bind (33.0%) (23).
Using our bound and free data sets, the performance results of all four servers are summarized in Table 2. Since DR_Bind does not aim to predict all residues at the protein–DNA interface, its sensitivity (35%) is lower than that of BINDN+ (45–48%), which has almost twice the number of predictions (i.e. TP + FP). Rather than knowing all residues that comprise the protein–DNA interface, most biologists would be interested in testing if the predicted residues do indeed bind DNA and therefore, a method’s precision, which reflects the fraction of predicted residues that are correct. Compared with the other methods, DR_Bind yields a ≥10% higher precision for both data sets. To assess if the difference in precision using DR_Bind and the other three methods is statistically significant, we randomly chose 40 and 25 protein structures from the bound and free data sets, respectively, and computed the precision obtained by each of the four servers; this was repeated 1000 times. The precision values obtained by DR_bind using the DNA-bound (0.38–0.56) and DNA-free structures (0.31–0.55) are generally higher than those obtained by the other three methods, as shown in Figure 2. This is also shown by the paired t-test, which was used to test the null hypothesis that DR_Bind does ‘not’ yield higher precision than the other three methods. The resulting P < 0.00001 for both bound and free data sets rejected the null hypothesis (Supplementary Table S3). Hence, an experimentalist would likely find more residues predicted by DR_bind to bind DNA compared with those predicted by sequence-based methods, thus saving time and costs.
Table 2.
Comparison of the performance measures of DR_Bind, BindN+, NAPS and DNABINDPROT using the same data set of 83 DNA bounda or 55 DNA-free protein structuresb,c
| Server | DR_Bind | BindN+ | NAPS | DNABINDPROT |
|---|---|---|---|---|
| TP | 728 (419) | 1013 (542) | 328 (180) | 244 (169) |
| FP | 831 (566) | 1798 (1129) | 733 (459) | 1040 (772) |
| TN | 18 128 (11 596) | 17 161 (11 033) | 18 226 (11 703) | 17 919 (11 390) |
| FN | 1362 (792) | 1077 (669) | 1762 (1031) | 1846 (1042) |
| Precision | 0.47 (0.43) | 0.36 (0.32) | 0.31 (0.28) | 0.19 (0.18) |
| Sensitivity | 0.35 (0.35) | 0.48 (0.45) | 0.16 (0.15) | 0.12 (0.14) |
| Specificity | 0.96 (0.95) | 0.91 (0.91) | 0.96 (0.96) | 0.95 (0.94) |
| Accuracy | 0.90 (0.90) | 0.86 (0.87) | 0.88 (0.89) | 0.86 (0.86) |
| mcc | 0.35 (0.33) | 0.34 (0.31) | 0.16 (0.15) | 0.08 (0.09) |
aThe PDB entries are listed in Supplementary Table S1; the total number of residues in the data set is 21 049, out of which 2090 residues are DNA-binding (=TP+FN) and 18 959 residues are non-DNA-binding (=FP+TN).
bPerformance measures based on the DNA-free protein structures are in the parentheses.
cThe PDB entries are listed in Supplementary Table S1; the total number of residues in the dataset is 13 373, out of which 1211 residues are DNA-binding (=TP+FN) and 12 162 residues are non-DNA-binding (=FP+TN).
Compared with sequence-based methods to predict DNA-binding residues, the structure-based DR_bind approach incorporates structural information (that is, electrostatics and geometry) of the query protein. Therefore, it would be expected to perform much better than sequence-based methods when evolutionary information for a query protein is not available. To show the importance of additional structural information, we tested the structure- and sequence-based methods on a set of 15 non-redundant DNA-bound protein structures with no or unreliable ConSurf conservation profiles. Note that DNABINDPROT could not be applied to this set of ‘unique’ DNA-binding proteins because it does not yield predictions for proteins without ConSurf-DB conservation data. The performance results of DR_bind, BINDN+ and NAPS in Table 3 show that the difference in performance between DR_bind and the two sequence-based methods become more apparent for proteins without conservation data: the precision of DR_bind (47%) is nearly twice that of BINDN+ (27%) and NAPS (23%). Thus, for DNA-binding proteins with no or insufficient homologs, DR_bind could provide a significantly higher fraction of correctly predicted DNA-binding residues than sequence-based methods.
Table 3.
Comparison of the performance measures of DR_Bind, BindN+ and NAPS using the same data set of 15 DNA-bound protein structures with no or insufficient close homologsa
| Server | DR_Bind | BindN+ | NAPS |
|---|---|---|---|
| TP | 110 | 230 | 34 |
| FP | 122 | 618 | 115 |
| TN | 2585 | 2089 | 2592 |
| FN | 292 | 172 | 368 |
| Precision | 0.47 | 0.27 | 0.23 |
| Sensitivity | 0.27 | 0.57 | 0.08 |
| Specificity | 0.95 | 0.77 | 0.96 |
| Accuracy | 0.87 | 0.75 | 0.84 |
| mcc | 0.29 | 0.26 | 0.07 |
aThe PDB entries are listed in Supplementary Table S2; the total number of residues in the data set is 3109, out of which 402 residues are DNA-binding (=TP+FN) and 2707 residues are non DNA-binding (=FP+TN).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–3.
FUNDING
Academia Sinica and the National Science Council, Taiwan. Funding for open access charge: National Science Council, Taiwan [NSC 95-2113-M-001-038-MY5] (to C.L.).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Karen Sargsyan for helpful discussion.
REFERENCES
- 1.Strong MJ, Volkening K, Hammond R, Yang W, Strong W, Leystra-Lantz C, Shoesmith C. TDP43 is a human low molecular weight neurofilament (hNFL) mRNA-binding protein. Mol. Cell. Neurosci. 2007;35:320–327. doi: 10.1016/j.mcn.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 2.Pavletich NP, Chambers KA, Pabo CO. The DNA-binding domain of p53 contains the four conserved regions and the major mutation hot spots. Genes Dev. 1993;7:2556–2564. doi: 10.1101/gad.7.12b.2556. [DOI] [PubMed] [Google Scholar]
- 3.Ahmad S, Gromiha MM, Sarai A. Analysis and prediction of DNA-binding proteins and their binding residues based on composition, sequence and structural information. Bioinformatics. 2004;20:477–486. doi: 10.1093/bioinformatics/btg432. [DOI] [PubMed] [Google Scholar]
- 4.Keil M, Exner TE, Brickmann J. Pattern recognition strategies for molecular surfaces: III. Binding site prediction with a neural network. J. Comput. Chem. 2004;25:779–789. doi: 10.1002/jcc.10361. [DOI] [PubMed] [Google Scholar]
- 5.Ahmad S, Sarai A. PSSM-based prediction of DNA binding sites in proteins. BMC Bioinformatics. 2005;6:33. doi: 10.1186/1471-2105-6-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yan C, Terribilini M, Wu F, Jernigan R, Dobbs D, Honavar V. Predicting DNA-binding sites of proteins from amino acid sequence. BMC Bioinformatics. 2006;7:262. doi: 10.1186/1471-2105-7-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuznetsov IB, Gou Z, Li R, Hwang S. Using evolutionary and structural information to predict DNA-binding sites on DNA-binding proteins. Proteins. 2006;64:19–27. doi: 10.1002/prot.20977. [DOI] [PubMed] [Google Scholar]
- 8.Hwang S, Gou Z, Kuznetsov IB. DP-Bind: a web server for sequence-based prediction of DNA-binding residues in DNA-binding proteins. Bioinformatics. 2007;23:634. doi: 10.1093/bioinformatics/btl672. [DOI] [PubMed] [Google Scholar]
- 9.Ofran Y, Mysore V, Rost B. Prediction of DNA-binding residues from sequence. Bioinformatics. 2007;23:i347–i353. doi: 10.1093/bioinformatics/btm174. [DOI] [PubMed] [Google Scholar]
- 10.Chu W, Huang Y, Huang C, Cheng Y, Huang C, Oyang Y. ProteDNA: a sequence-based predictor of sequence-specific DNA-binding residues in transcription factors. Nucleic Acids Res. 2009;37:W396–W401. doi: 10.1093/nar/gkp449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang L, Brown SJ. BindN: a web-based tool for efficient prediction of DNA and RNA binding sites in amino acid sequences. Nucleic Acids Res. 2006;34:W243–W248. doi: 10.1093/nar/gkl298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang L, Huang C, Yang M, Yang J. BindN+ for accurate prediction of DNA and RNA-binding residues from protein sequence features. BMC Syst. Biol. 2010;4:S3. doi: 10.1186/1752-0509-4-S1-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu J, Liu H, Duan X, Ding Y, Wu H, Bai Y, Sun X. Prediction of DNA-binding residues in proteins from amino acid sequences using a random forest model with a hybrid feature. Bioinformatics. 2009;25:30–35. doi: 10.1093/bioinformatics/btn583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang L, Yang MQ, Yang JY. Prediction of DNA-binding residues from protein sequence information using random forest. BMC Genomics. 2009;10:S1. doi: 10.1186/1471-2164-10-S1-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carson MB, Langlois R, Lu H. NAPS: a residue-level nucleic acid-binding prediction server. Nucleic Acids Res. 2010;38:W431–W435. doi: 10.1093/nar/gkq361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Si J, Zhang Z, Lin B, Schroeder M, Huang B. MetaDBSite: a meta approach to improve protein DNA-binding sites prediction. BMC Syst. Biol. 2011;5:S7. doi: 10.1186/1752-0509-5-S1-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jones S, Shanahan HP, Berman HM, Thornton JM. Using electrostatic potentials to predict DNA-binding sites on DNA-binding proteins. Nucleic Acids Res. 2003;31:7189–7198. doi: 10.1093/nar/gkg922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stawiski EW, Gregoret LM, Mandel-Gutfreund Y. Annotating nucleic acid-binding function based on protein structure. J. Mol. Biol. 2003;326:1065–1079. doi: 10.1016/s0022-2836(03)00031-7. [DOI] [PubMed] [Google Scholar]
- 19.Tsuchiya Y, Kinoshita K, Nakamura H. Structure-based prediction of DNA-binding sites on proteins using the empirical preference of electrostatic potential and the shape of molecular surfaces. Proteins. 2004;55:885–894. doi: 10.1002/prot.20111. [DOI] [PubMed] [Google Scholar]
- 20.Shanahan HP, Garcia MA, Jones S, Thornton JM. Identifying DNA-binding proteins using structural motifs and the electrostatic potential. Nucleic Acids Res. 2004;32:4732–4741. doi: 10.1093/nar/gkh803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ferrer-Costa C, Shanahan HP, Jones S, Thornton JM. HTHquery: a method for detecting DNA-binding proteins with a helix-turn-helix structural motif. Bioinformatics. 2005;21:3679–3680. doi: 10.1093/bioinformatics/bti575. [DOI] [PubMed] [Google Scholar]
- 22.Wu CY, Chen YC, Lim C. A structural-alphabet-based strategy for finding structural motifs across protein families. Nucleic Acids Res. 2010;38:e150. doi: 10.1093/nar/gkq478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ozbek P, Soner S, Erman B, Haliloglu T. DNABINDPROT: fluctuation-based predictor of DNA-binding residues within a network of interacting residues. Nucleic Acids Res. 2010;38:W417–W423. doi: 10.1093/nar/gkq396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsuchiya Y, Kinoshita K, Nakamura H. PreDs: a server for predicting dsDNA-binding site on protein molecular surfaces. Bioinformatics. 2005;21:1721–1723. doi: 10.1093/bioinformatics/bti232. [DOI] [PubMed] [Google Scholar]
- 25.Tjong H, Zhou HX. DISPLAR: an accurate method for predicting DNA-binding sites on protein surfaces. Nucleic Acids Res. 2007;35:1465. doi: 10.1093/nar/gkm008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gao M, Skolnick J. DBD-Hunter: a knowledge-based method for the prediction of DNA-protein interactions. Nucleic Acids Res. 2008;36:3978–3992. doi: 10.1093/nar/gkn332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen YC, Wu CY, Lim C. Predicting DNA-binding sites on proteins from electrostatic stabilization upon mutation to Asp/Glu and evolutionary conservation. Proteins. 2007;67:671–680. doi: 10.1002/prot.21366. [DOI] [PubMed] [Google Scholar]
- 28.van Dijk M, Bonvin AMJJ. A protein–DNA docking benchmark. Nucleic Acids Res. 2008;36:e88. doi: 10.1093/nar/gkn386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen YC, Lim C. Common physical basis of macromolecule-binding sites in proteins. Nucleic Acids Res. 2008;36:7078–7087. doi: 10.1093/nar/gkn868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Berman HM, Battistuz T, Bhat TN, Bluhm WF, Bourne PE, Burkhardt K, Iype L, Jain S, Fagan P, Marvin J, et al. The Protein Data Bank. Acta Crystallogr. D. 2002;58:899–907. doi: 10.1107/s0907444902003451. [DOI] [PubMed] [Google Scholar]
- 31.Pearl F, Todd A, Sillitoe I, Dibley M, Redfern O, Lewis T, Bennett C, Marsden R, Grant A, Lee D, et al. The CATH Domain Structure Database and related resources Gene3D and DHS provide comprehensive domain family information for genome analysis. Nucleic Acids Res. 2005;33:D247–D251. doi: 10.1093/nar/gki024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goldenberg O, Erez E, Nimrod G, Ben-Tal N. The ConSurf-DB: pre-calculated evolutionary conservation profiles of protein structures. Nucleic Acids Res. 2009;37:D323–D327. doi: 10.1093/nar/gkn822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taylor WR, Orengo CA. Protein structure alignment. J. Mol. Biol. 1989;208:1–22. doi: 10.1016/0022-2836(89)90084-3. [DOI] [PubMed] [Google Scholar]
- 34.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignments through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McDonald IK, Thornton JM. Satisfying hydrogen bonding potential in proteins. J. Mol. Biol. 1994;238:777–793. doi: 10.1006/jmbi.1994.1334. [DOI] [PubMed] [Google Scholar]
- 36.Miller S, Janin J, Lesk AM, Chothia C. Interior and surface of monomeric proteins. J. Mol. Biol. 1987;196:641–656. doi: 10.1016/0022-2836(87)90038-6. [DOI] [PubMed] [Google Scholar]
- 37.Koradi R, Billeter M, Wuthrich K. MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graph. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 38.Canutescu AA, Shelenkov AA, Dunbrack RL., Jr A graph-theory algorithm for rapid protein side-chain prediction. Protein Sci. 2003;12:2001–2014. doi: 10.1110/ps.03154503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Case DA, Cheatham TE, III, Darden T, Gohlke H, Luo R, Merz KM, Jr, Onufriev A, Simmerling C, Wang B, Woods RJ. The Amber biomolecular simulation programs. J. Comput. Chem. 2005;26:1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Duan Y, Wu C, Chowdhury S, Lee MC, Xiong G, Zhang W, Yang R, Cieplak P, Luo R, Lee T, et al. A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J. Comput. Chem. 2003;24:1999–2012. doi: 10.1002/jcc.10349. [DOI] [PubMed] [Google Scholar]
- 41.Ashkenazy H, Erez E, Martz E, Pupko T, Ben-Tal N. ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 2010;38:W529–W533. doi: 10.1093/nar/gkq399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Landau M, Mayrose I, Rosenberg Y, Glaser F, Martz E, Pupko T, Ben-Tal N. ConSurf 2005: the projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res. 2005;33:299–302. doi: 10.1093/nar/gki370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Glaser F, Pupko T, Paz I, Bell RE, Bechor-Shental D, Martz E, Ben-Tal N. ConSurf: identification of functional regions in proteins by surface-mapping of phylogenetic information. Bioinformatics. 2003;19:163–164. doi: 10.1093/bioinformatics/19.1.163. [DOI] [PubMed] [Google Scholar]
- 44.Chen YC, Lim C. Predicting RNA-binding sites from the protein structure based on electrostatics, evolution and geometry. Nucleic Acids Res. 2008;36:e29. doi: 10.1093/nar/gkn008. [DOI] [PMC free article] [PubMed] [Google Scholar]