Abstract
Analyses of multiple sequence alignments generally focus on well-defined conserved sequence blocks, while the rest of the alignment is largely ignored or discarded. This is especially true in phylogenomics, where large multigene datasets are produced through automated pipelines. However, some of the most powerful phylogenetic markers have been found in the variable length regions of multiple alignments, particularly insertions/deletions (indels) in protein sequences. We have developed Sequence Feature and Indel Region Extractor (SeqFIRE) to enable the automated identification and extraction of indels from protein sequence alignments. The program can also extract conserved blocks and identify fast evolving sites using a combination of conservation and entropy. All major variables can be adjusted by the user, allowing them to identify the sets of variables most suited to a particular analysis or dataset. Thus, all major tasks in preparing an alignment for further analysis are combined in a single flexible and user-friendly program. The output includes a numbered list of indels, alignments in NEXUS format with indels annotated or removed and indel-only matrices. SeqFIRE is a user-friendly web application, freely available online at www.seqfire.org/.
INTRODUCTION
Multiple sequence alignment (MSA) is a core bioinformatic tool with many different applications (1). Most of these applications focus on the well-conserved blocks of the MSA where alignment among the sequences is unambiguous. Regions that vary in length among the sequences, the so-called gapped or insertion/deletion (indel) regions, are more generally discarded. This is especially true in phylogenetics, where indel regions are usually avoided because of their uncertain homology or because of the theoretical complexity of weighting indels, which regardless of size may still represent a single evolutionary event (2). This wholesale discarding of indel information is unfortunate, as it has been recognized for some time that rare genomic changes such as indels are a unique and potentially very powerful class of phylogenetic marker (3).
The phylogenetic power of indels stems from the fact that, in contrast to single amino acid or nucleotide substitutions, indels are (i) less prone to homoplasy (multiple independent origins) because they are more complex, (ii) more stable because they are difficult to fully reverse and (iii) easier to assess for homology, particularly when they cover multiple alignment columns (3). A number of important evolutionary discoveries have relied heavily on indels such as recognition of the eukaryotic supergroup Opisthokonta (Holozoa + Holomycota) (4), rooting the tree of eutherian mammals (5) and supporting the possible eocyte origin of eukaryotes (6–7). Nonetheless, the potential of indels as phylogenetic markers is generally wasted, particularly with the increasing emphasis on large multigene phylogenies. These large datasets are, generally by necessity assembled by pipelines that automatically discard regions considered unsuitable for phylogenetic tree reconstruction (8). Thus, despite the explosion in molecular data and molecular phylogenetic dataset size, indel information is being largely lost.
We developed Sequence Feature and Indel Region Extractor (SeqFIRE) to facilitate automated and systematic evaluation and extraction of indel regions in MSAs. The program also performs the more standard extraction of conserved blocks for use in phylogenetic analysis. Thus, the program performs all major tasks in preparing an MSA for further analysis. SeqFIRE is designed so that the user can easily adjust all major parameters, which makes the program more flexible than other currently available alignment editors (9,10). This allows the user to select optimal parameters for a particular dataset or to experiment with a range of parameters in order to examine different possible interpretations of potentially important indel regions. Visualization of alignments is implemented through Jalview (11), including annotation of conserved block and indel regions. SeqFIRE is open-source software and is platform independent. A stand-alone version is also provided for pipelining or running locally. The SeqFIRE source code is available from the program web site (www.seqfire.org).
INDELS AND INDEL REGIONS
For our purposes here, we define MSAs as comprising two types of regions: conserved blocks and insertion/deletion (indel) regions. Conserved blocks are alignable without gaps across all sequences and are inferred to be homologous throughout their length (1). These regions are relatively easy to define and to work with and are generally useful for phylogenetic tree reconstruction. In contrast, indel regions show a range of lengths among the sequences. These regions vary from easy to extremely difficult to define, depending on the complexity of the indel and the degree of sequence conservation in the surrounding alignment (12).
We further recognize two types of indels here, simple and complex. Simple indels are defined as those that occur in only two states, that is, the indel is either present or absent. Such indels appear to represent a single evolutionary event. All other indels are classified here as complex indels. These are gapped regions that exist in three or more states and therefore result from two or more evolutionary events occurring in the same or over-lapping regions. The interpretation of indels is further complicated by the fact that they tend to occur in alignment regions of low sequence conservation and also tend to be rapidly evolving themselves (12). All of these factors need to be considered in order to evaluate the placement of an indel within an MSA and the number of events that have contributed to the indel itself.
Thus, there are two main components to interpreting an indel region: the boundaries of the region and the number of indel events that have occurred within it. Since it is not always possible to know which solution is ‘correct’, SeqFIRE uses a conservative approach to the problem of defining indel boundaries by working with ‘indel regions’. These are defined as a set of adjacent gap-containing alignment columns plus all flanking non-gapped columns with sequence conservation below a designated threshold (default or user-defined). The user can then adjust the parameters used in defining these indel regions in order to examine a range of possible interpretations.
THE SeqFIRE PROGRAM
The SeqFIRE core program is implemented in Python, and the web interface uses PHP and HTML. The program consists of two modules. These are an indel region module for identification and extraction of indels, with or without surrounding regions of ambiguous alignment, and a conserved block module for identification and extraction of conserved alignment blocks.
Input
SeqFIRE uses aligned protein sequences in FASTA format as input. Single MSA input files can be uploaded or pasted directly into an input box. For batch analysis, the individual MSA input files must first be merged into a single large (multiple MSA) input file. This can be done using SeqFIREprep, a small stand-alone program that can be downloaded from the web site. SeqFIREprep can also be used after the analysis, to split the program output back into individual alignment-specific files.
Algorithms used in the indel region module
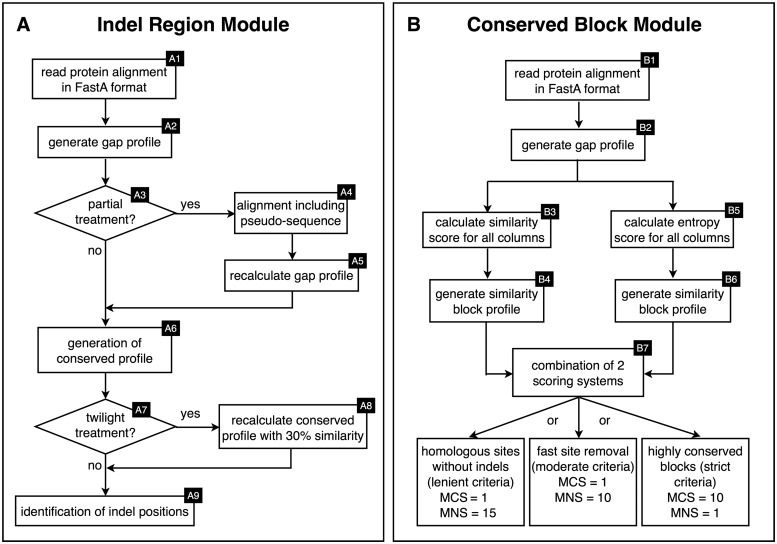
The indel region module functions in the identification, classification and extraction of indel regions from MSAs (Figure 1A). The process begins with the generation of a gap profile, which is a single string containing scores for every alignment column. As a result, any column with a gap in any sequence is scored as a ‘gap column’ and all other columns are scored as gap-free (Steps A1 and A2, Figure 1).
Figure 1.
Work flow for SeqFIRE, a user-friendly web application for automated identification and extraction of indels and conserved blocks from MSAs. The workflow for the (A) indel and (B) conserved block modules of SeqFIRE are shown on the left and right, respectively. Boxes indicate processes and diamonds indicate suggested parameters for specific steps. Numbers in the upper right-hand corners of boxes indicate different steps in the process as described in the text. For the conserved block module, MNS refers to minimum non-conserved site threshold and MCS to minimum conserved site threshold.
This scoring can be problematic if there are incomplete sequences in the MSA, as these will give rise to large gapped regions in the profile, most commonly at the beginning or end of an alignment. This will result in the masking of any other possibly useful information in these regions. SeqFIRE allows the user to select a partial treatment option for an MSA with incomplete sequences. This treatment fills in large terminal gaps with a pseudo-sequence before the gap profile is generated (Steps A3–A5, Figure 1). The process begins by designating any sequence with continuously missing data for over 60% (default) of an end-terminal region as a ‘designated partial sequence’ (DPS). DPSs are then modified as follows (using default = 60%): positions that are missing in the DPS but present in ≥60% of the remaining sequences are designated as unknown (‘?’) and positions missing in the DPS that are present in <60% of the other sequences are designated as gaps.
Once the gap profile is generated, all gap-free positions are assigned a similarity score. This uses similarity groups based on a user-selected substitution matrix (PAM60, PAM250, BLOSUM40, BLOSUM62 or BLOSUM80; default = NONE) (Steps A6–A8, Figure 1; Supplementary Material S1). For each non-gap column, the number of amino acids for each similarity group is then counted. If any of these counts are above the selected threshold (default = 75%), the site will be classified as a ‘conserved position’. Any column with a similarity score below the threshold will be classed as a ‘divergent position’. Since homologous proteins with sequence similarity as low as 25–35% can still have the same or similar structure (13), SeqFIRE provides a ‘twilight treatment’ option, which automatically sets the similarity threshold to 30%. If the default option (NONE) is used, only identical residues will be counted towards the similarity score.
SeqFIRE uses the indel profile to systematically extract all indel regions from the MSA beginning at its amino terminus. The ‘minimum residue value’ (default = 3) defines the minimum number of contiguous, conserved columns in the MSA that are required to flank or ‘anchor’ an indel region. This has the result that any highly variable columns adjacent to gap columns will also be included in the indel region. As explained above, this is because an indel can often be extended into such regions with little, if any decrease in alignment quality score. The minimum residue value also prevents an indel from being split due to the presence of one or a few gap-free alignment columns within an indel region.
Algorithms used in the conserved block module
In addition to extracting indels, SeqFIRE can also output the non-indel portions of an MSA with varying user-selected levels of stringency. These are designated low, moderate or high. At low stringency, the program will output all alignment blocks between the indel regions, including the three conserved residues flanking each indel. This is essentially the alignment with all gap regions removed. At moderate stringency, the program will further clean the alignment by removing fast evolving positions as defined by a combination of entropy and similarity scores (Supplementary Material S1). This is similar to the phylogenetic practice of ‘fast site removal’ (14,15). At high stringency, SeqFIRE will remove all but the most highly conserved alignment blocks. This function can be used to identify universal sequence motifs, which can be useful for applications such as polymerase chain reaction primer design or diagnostics.
The flow of the conserved block module is shown in Figure 1B and described generally here and in detail in Supplementary Table S1. As with the indel module, all decisions are based on a gap profile. The calculation starts by recording all positions where a gap is present in a designated percentage of all sequences (default = 40%). The remaining (non-gap) sites are then assigned two scores, a similarity score and an entropy score. The similarity score is calculated as described above for the indel module, and then trimmed of isolated conserved or non-conserved alignment columns by applying separate minimum size limits for non-conserved and conserved blocks (default = 3 and 1, respectively) (Supplementary Material S1). The entropy profile is generated using Shannon entropy (H), where higher values indicate a greater diversity of residues at a given alignment position. The similarity and entropy profiles are then combined either by union or intersection, depending on whether the user selects strict or relaxed criteria, respectively. This combined profile is then used to identify the final set of conserved blocks (Supplementary Material S1).
Output
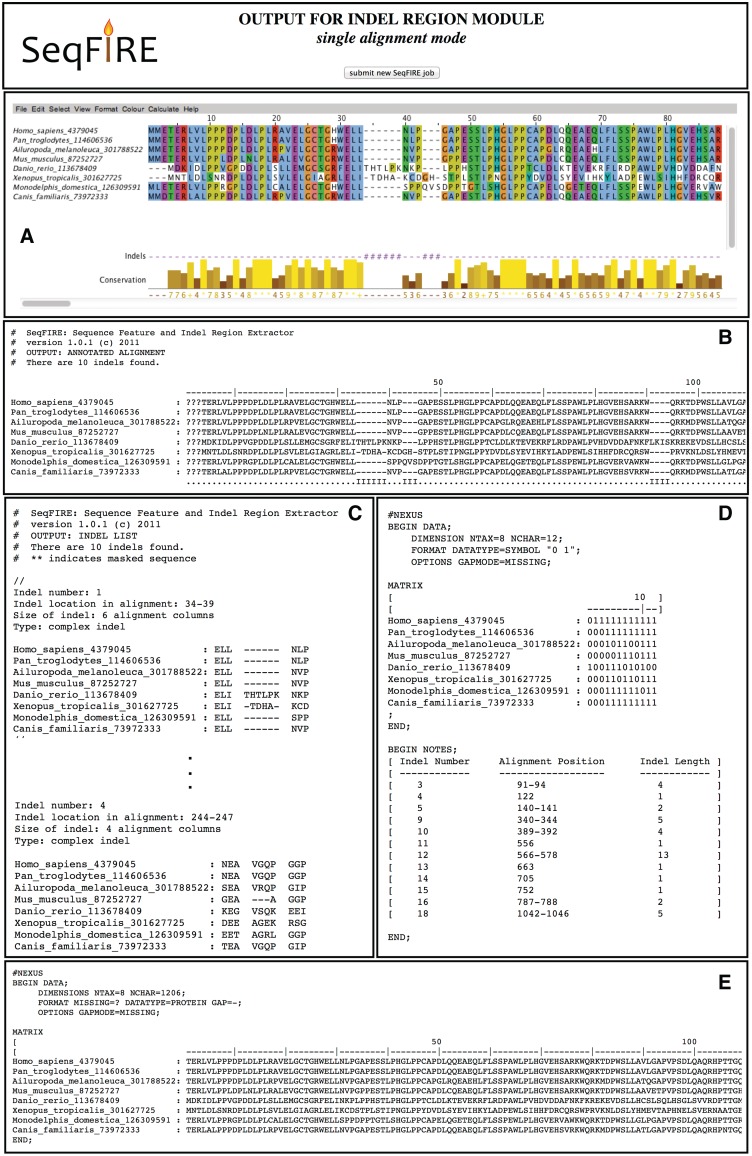
The output for the indel module (Figure 2) consists of
Annotated alignment in Jalview
Annotated alignment in text mode
Indel list
Indel matrix
Masked alignment
The annotated alignment consists of the MSA with the indel profile displayed below it. The indel list is a sequentially numbered list of all indels. The indel matrix is a presence/absence matrix in NEXUS format for the complete set of simple (two state) indels. The masked alignment is the MSA with indel regions removed.
Figure 2.
Example output from the SeqFIRE indel module. The module produces five different outputs (A–E). The alignment with indel annotation is visualized in Jalview (A) and text mode (B). The indels list is a numbered sequential list of all indels including the location of the indel in the alignment and the full sequence of the indel region for all taxa (C). The simple indel matrix is a NEXUS-formatted matrix with all simple indels scored as 0 or 1 (absence or presence) for all taxa (D). The indel module also outputs an alignment with all indel regions removed, also in NEXUS format (E). Outputs B–E can be downloaded as a single file or separately using links at the top of the output page.
The output for the conserved block module consists of
Annotated alignment in Jalview
Annotated alignment in text mode
Full alignment plus indel profile in FastA format
Masked alignment (indel regions deleted) in FastA format
Full alignment with indels listed in a NEXUS ‘character block’
Masked alignment in NEXUS format
All output is in NEXUS format. For the full alignment plus indel profile, the profile is enclosed in hard brackets (‘[ ]’) so as not to interfere in phylogenetic analysis. The full alignment with indels listed in a character block allows the user to delete these regions from a phylogenetic analysis using the NEXUS delete character command (‘del charset’). The masked alignment plus indel matrix allows the user to use the indels as additional phylogenetic characters. Jalview is also used on the web site for visualization of the alignment with indel and conserved block profiles.
A performance test of the SeqFIRE conserved block module
We compared SeqFIRE’s conserved block module with GBlocks (9,10), currently the most widely used publicly available program for conserved block identification. Comparisons were run using three different reference levels of BAliBASE 3.0 (16), a benchmark database for sequence alignment methods and tools. Five alignments were selected at random from each reference level, which represent different levels and types of sequence conservation (Table 1). The reference 1 V1 subset consists of alignments with <20% sequence similarity, including large internal insertions (>35 residues). Alignments in the reference 1 V2 subset share 20–40% similarity more or less equally among all sequences. Reference 3 alignments include several protein subfamilies within the same alignment, so that these share >40% similarity within the same subfamily but <20% similarity between the different subfamilies.
Table 1.
Performance of SeqFIRE and GBlocks in detecting conserved blocks within BAliBASE (16) benchmark alignments
| Test alignment | Original alignment (sites) | GBlocks |
SeqFIRE |
|||
|---|---|---|---|---|---|---|
| Less stringency | More stringency | Low stringency | Medium stringency | High stringency | ||
| Ref 1 V1 (<20% similarity) | ||||||
| BB1103 | 582 | 162 (27.8%) | 42 (7.2%) | 218 (37.5%) | 215 (36.9%) | 49 (8.4%) |
| BB1105 | 609 | 14 (2.3%) | 0 (0.0%) | 336 (55.2%) | 314 (51.6%) | 0 (0.0%) |
| BB1106 | 385 | 29 (7.5%) | 0 (0.0%) | 205 (53.2%) | 193 (50.1%) | 0 (0.0%) |
| BB11031 | 882 | 26 (2.6%) | 0 (0.0%) | 278 (31.5%) | 253 (28.7%) | 6 (0.7%) |
| BB11036 | 525 | 69 (13.1%) | 0 (0.0%) | 322 (61.3%) | 304 (57.9%) | 39 (7.4%) |
| Ref 1 V2 (20–40% similarity) | ||||||
| BB12001 | 623 | 193 (31.0%) | 83 (13.3%) | 372 (59.7%) | 361 (57.9%) | 107 (17.2%) |
| BB12004 | 312 | 152 (48.7%) | 40 (12.8%) | 226 (72.4%) | 226 (72.4%) | 92 (29.5%) |
| BB12017 | 586 | 318 (54.3%) | 229 (39.1%) | 425 (72.5%) | 414 (70.6%) | 233 (39.8%) |
| BB12030 | 1247 | 279 (22.4%) | 83 (6.7%) | 738 (59.2%) | 738 (59.2%) | 192 (15.4%) |
| BB12043 | 786 | 120 (15.3%) | 13 (1.7%) | 211 (26.8%) | 210 (26.7%) | 91 (11.6%) |
| Ref 3 (>40% similarity) | ||||||
| BB30008 | 1413 | 158 (11.2%) | 28 (2.0%) | 333 (23.6%) | 323 (22.9%) | 93 (6.6%) |
| BB30009 | 278 | 48 (17.3%) | 0 (0.0%) | 184 (66.2%) | 155 (55.8%) | 5 (1.8%) |
| BB30021 | 631 | 25 (4.0%) | 0 (0.0%) | 151 (23.9%) | 131 (20.8%) | 12 (1.9%) |
| BB30027 | 239 | 49 (20.5%) | 0 (0.0%) | 67 (28.0%) | 60 (25.1%) | 5 (2.1%) |
| BB30030 | 2015 | 129 (6.4%) | 21 (1.0%) | 254 (12.6%) | 236 (11.7%) | 39 (1.9%) |
Test alignment numbers refer to BAliBASE accession numbers for three different levels of sequence conservation: Ref 1V1, Ref 1V2 and Ref 3. GBlocks was tested at the two stringency levels provided by the web server, while SeqFIRE was tested at three levels using a combination of user-defined options (for details see text). For each stringency level, the number of conserved positions is listed with the percentage of retained sites shown below in parentheses.
SeqFIRE was tested at three different stringency levels, designated here as low, medium and high. For low stringency, the parameters consisted of 40% accept gaps, 55% amino acid conservation threshold, minimum conserved block size of one and maximum non-conserved block size of 15, with the block profiles combined using the union method. For medium stringency, the first three parameters were re-set to 35% accept gaps, 65% amino acid conservation threshold and minimum conserved block size of 3, with the remaining parameters unchanged. The high stringency condition used the same parameters as the medium run except the amino acid conservation threshold was increased to 75% and the intersection method was used to combine the profiles. GBlocks was run at lower and higher stringency using the web server version (http://molevol.cmima.csic.es/castresana/Gblocks_server.html). For less stringency, all default options were selected. For high stringency running, the option ‘do not allow many contiguous nonconserved positions’ was selected.
The comparative performance tests show that SeqFIRE and GBlocks give fairly similar results for high stringency conditions, although SeqFIRE consistently retains more alignment sites than GBlocks (Table 1), including some apparently quite well-conserved patches (Figure 3). Meanwhile, the single less stringent option available through the GBlocks web server gives results that are intermediate between the high and medium stringency levels used here for SeqFIRE. This tends to result in at least twice as many alignment columns identified as potentially homologous by SeqFIRE than by GBlocks, and sometimes considerably more than that (Table 1). Thus, SeqFIRE gives the user the option to consider many more alignment positions for further analysis or to adjust the program variables to gradually increase the stringency of selection to an appropriate level as judged by visual inspection of the alignment mask in JalView. Once set, these variables can then be implemented in an automated manner for groups of alignments aimed at a similar phylogenetic depth. It should be noted that the lowest recommended stringency level used here for SeqFIRE finds a few additional sites, particularly for the low (Ref 1 V1, Table 1) and mixed conservation alignments (Ref 3, Table 1). The fact that there is not a large increase between the moderate and low stringency levels suggests that the program is still capable of screening out spurious alignment positions even at low stringency.
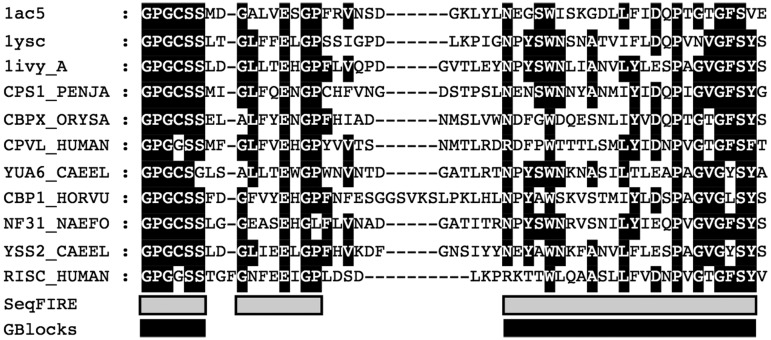
Figure 3.
Example of SeqFIRE and GBlocks detection of conserved alignment regions under high stringency criteria. A fragment of BAliBASE reference 1 V2 alignment number BB12001 is shown between positions 129 and 187. The gray bars below the alignment indicate the conserved blocks detected by SeqFIRE and the black bars show the conserved blocks detected by GBlocks. The dark background within the alignment indicates conserved amino acids.
GBlocks was designed to be a conservative program, erring on the side of caution in identifying conserved alignment blocks (9). This is a safe and useful strategy, particularly when alignments are used for examining deep phylogenetic nodes, such as those on which the program was benchmarked (9). However, this can mean that potentially phylogenetically useful information is lost, particularly for less conserved proteins being used to examine more shallow evolutionary nodes. The main strength of SeqFIRE in identifying conserved blocks is that it allows the user to decide the level of stringency appropriate for their particular dataset and phylogenetic question, which can vary widely. Most importantly, since the user-defined variables are clearly specified and then implemented automatically by the program, alignment site selection is still done in a transparent and reproducible manner.
CONCLUSION
Nearly all MSAs require some ‘editing’ to remove regions with gaps and/or uncertain alignment, especially if the alignment is to be used as input for phylogenetic analysis. The traditional and simplest way of doing this editing is to remove all alignment columns with gaps in any sequence (9,17–19). This ignores potential ambiguity in the exact placement of an indel within an alignment as well as the loss of information when incomplete sequences are present. More sophisticated MSA editing applications overcome these problems by using consensus sequences to define conserved alignment blocks (9,10,20). However, these programs still universally focus on defining conserved alignment blocks. Currently available programs also tend to use strict criteria that allow for little, if any user input. Most importantly, none of these programs assesses the phylogenetic potential of indels.
SeqFIRE was developed with the primary purpose of allowing users to explore and extract indel regions from MSAs. A module for extracting conserved blocks is also included in order to provide a complete sequence editing service. The aim is to allow indel assessment to become a routine part of any molecular phylogenetic analysis. The program includes an easy-to-use web interface and a stand-alone version that can be used to pipeline large amounts of data, such as for multigene phylogenies (21–23). The program allows users to select from a range of variables for all major parameters used in the analysis and to easily adjust these parameters in order to optimize them for a particular dataset or to explore alternative interpretations of the data. This is especially important for indels, because defining indels is often not straightforward, even for indels that may ultimately prove to be phylogenetically informative (4).
There are currently three indel databases widely available—Indel PDB (24), IndelFR (25) and INDELSCAN (26). These each use slightly different approaches to identifying indel regions. Indel PDB uses protein sequences aligned by BLASTp (24). Its aim is to examine the placement of indels within protein structures without attention to indel boundaries or evolutionary patterns. INDELSCAN (26) is a DNA indel database that uses pairwise alignments plus one or more outgroup sequences. Again, the indel is defined purely as a region of continuous gaps between the two ingroup sequences, and outgroup sequences are used only to classify gaps as insertions or deletions. IndelFR (25) uses a pairwise structure alignment program, PDBeFold (27) and extracts the regions of the alignment immediately bordering indels (regions of continuous gap in the alignment). Thus, all three currently available indel databases use pairwise alignments and define indels as any gap-containing region in the alignment. SeqFIRE differs substantially from these by extracting indels from protein MSAs. Thus, only SeqFIRE can distinguish simple from complex indels, as any indel appears simple in a pairwise alignment. In addition, SeqFIRE is the only current indel-extracting program that considers the quality of the indel flanking regions.
Indels are potentially very powerful phylogenetic characters either used alone as individual markers (5–7,28,29) or combined with other data in a mixed-data phylogenetic analysis. However, simply leaving an indel in an alignment used for phylogenetic analysis is not justified, even for simple indels, as each gap column will be treated as a separate character. Thus, an indel will be automatically afforded a weight proportional to its length, for which there is no theoretical or empirical justification. Nonetheless, the problem of how to weight indels in phylogenetic reconstruction is a complex issue (30–32). Although various schemes have been proposed for weighting sequence indels (33–35), these are largely theoretical. Thus, an additional goal in developing SeqFIRE is to make it easier to preserve the indel information potentially available in large-scale phylogenomic studies. Such information can then be used to develop more realistic schemes for indel weighting based on how these characters behave over time.
Nearly all methods currently available for scoring indels in MSAs deal exclusively with DNA sequences (35,36). The simplest method proposed is to designate all gaps as a fifth character state (35). However, this method is problematic in the case of complex gaps for the reasons discussed above. Other DNA indel coding methods attempt to use complex indels by separating them into smaller simple indels, which are then scored as present/absent (35,36). This includes programs such as SeqState (37). However, breaking complex indels down into single events is difficult to do with accuracy and therefore can have a negative effect on the accuracy of tree building. The only other method currently available for scoring indels in protein sequence alignments is the program GapCoder (33). This uses a similar method to SeqFIRE, by scoring simple protein indels in a presence/absence matrix. Neither SeqFIRE nor GapCoder attempts to score complex indels. However, SeqFIRE goes further by extracting complex indels and designating them as such. This allows the user to examine and experiment with these potentially useful characters in order to make an informed assessment as to whether or not they might have further utility.
SeqFIRE is easy to use as a stand-alone program or to add as a pipeline to other processes. The program is written with standard Python modules, so the user does not need to deal with any Python dependencies. SeqFIRE is also useful as an educational tool, to help students visualize how different alignment parameters impact on indel and conserved block identification. Future plans for the program include pipelining existing and publicly available alignment programs. This will allow the user to begin with unaligned sequences or to re-align designated portions of existing MSAs in order to more fully explore the ‘alignment space’ surrounding individual indel regions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table 1 and Supplementary Material 1.
FUNDING
Royal Thai Government Scholarship and a graduate student fellowship from Uppsala University (to P.A.); Estonian Science Foundation Mobilitas [MJD99 and GLOTI9020 to G.C.A.]; the Center of Excellence in Chemical Biology, University of Tartu, Estonia (to G.C.A.) and the Swedish Research Council [2010-2771] (to S.L.B.). Funding for open access charge: Swedish Research Council [2010-2771] to senior author (S.L.B.).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank Anders Larsson for technical support in the construction of the SeqFIRE web server. They thank Allison Perrigo, Chen-jie Fu and Mikael Thollesson for helpful comments on the manuscript and members of the Systematic Biology Programme for helping to troubleshoot earlier versions of the program.
REFERENCES
- 1.Aniba MR, Poch O, Thompson JD. Issues in bioinformatics benchmarking: the case study of multiple sequence alignment. Nucleic Acids Res. 2010;38:7353–7363. doi: 10.1093/nar/gkq625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lockwood CA. Adaptation and functional integration in primate phylogenetics. J. Hum. Evol. 2007;52:490–503. doi: 10.1016/j.jhevol.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 3.Rokas A, Holland PWH. Rare genomic changes as a tool for phylogenetics. Trends Ecol. Evol. 2000;15:454–459. doi: 10.1016/s0169-5347(00)01967-4. [DOI] [PubMed] [Google Scholar]
- 4.Baldauf SL. A search for the origins of animals and fungi: comparing and combining molecular data. Am. Nat. 1999;154:178–188. doi: 10.1086/303292. [DOI] [PubMed] [Google Scholar]
- 5.de Jong WW, van Dijk MAM, Poux C, Kappé G, van Rheede T, Madsen O. Indels in protein-coding sequences of Euarchontoglires constrain the rooting of the eutherian tree. Mol. Phylogenet. Evol. 2003;28:328–340. doi: 10.1016/s1055-7903(03)00116-7. [DOI] [PubMed] [Google Scholar]
- 6.Rivera MC, Lake JA. Evidence that eukaryotes and eocyte prokaryotes are immediate relatives. Science. 1992;257:74–76. doi: 10.1126/science.1621096. [DOI] [PubMed] [Google Scholar]
- 7.Cox CJ, Foster PG, Hirt RP, Harris SR, Embley TM. The archaebacterial origin of eukaryotes. Proc. Natl Acad. Sci. USA. 2008;105:20356–20361. doi: 10.1073/pnas.0810647105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ciccarelli FD, Doerks T, von Mering C, Creevey CJ, Snel B, Bork P. Toward automatic reconstruction of a highly resolved tree of life. Science. 2006;311:1283–1287. doi: 10.1126/science.1123061. [DOI] [PubMed] [Google Scholar]
- 9.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- 10.Talavera G, Castresana J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 2007;56:564–577. doi: 10.1080/10635150701472164. [DOI] [PubMed] [Google Scholar]
- 11.Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. Jalview version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thorne JL. Models of protein sequence evolution and their applications. Curr. Opin. Genet. Dev. 2000;10:602–605. doi: 10.1016/s0959-437x(00)00142-8. [DOI] [PubMed] [Google Scholar]
- 13.Rost B. Twilight zone of protein sequence alignments. Protein Eng. 1999;12:85–94. doi: 10.1093/protein/12.2.85. [DOI] [PubMed] [Google Scholar]
- 14.Kumar S, Skjæveland Å, Orr RJS, Enger P, Ruden T, Mevik BH, Burki F, Botnen A, Shalchian-Tabrizi K. AIR: a batch-oriented web program package for construction of supermatrices ready for phylogenomic analyses. BMC Bioinformatics. 2009;10:357. doi: 10.1186/1471-2105-10-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hirt RP, Logsdon JM, Jr, Healy B, Dorey MW, Doolittle WF, Embley TM. Microsporidia are related to fungi: evidence from the largest subunit of RNA polymerase II and other proteins. Proc. Natl Acad. Sci. USA. 1999;96:580–585. doi: 10.1073/pnas.96.2.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thompson JD, Koehl P, Ripp R, Poch O. BAliBASE 3.0: latest developments of the multiple sequence alignment benchmark. Proteins. 2005;61:127–136. doi: 10.1002/prot.20527. [DOI] [PubMed] [Google Scholar]
- 17.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Löytynoja A, Goldman N. Phylogeny-aware gap placement prevents errors in sequence alignment and evolutionary analysis. Science. 2008;320:1632–1635. doi: 10.1126/science.1158395. [DOI] [PubMed] [Google Scholar]
- 19.Wu M, Chatterji S, Eisen JA. Accounting for alignment uncertainty in phylogenomics. PLoS One. 2012;7:e30288. doi: 10.1371/journal.pone.0030288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smagala JA, Dawson ED, Mehlmann M, Townsend MB, Kuchta RD, Rowlen KL. ConFind: a robust tool for conserved sequence identification. Bioinformatics. 2005;21:4420–4422. doi: 10.1093/bioinformatics/bti719. [DOI] [PubMed] [Google Scholar]
- 21.Dunn CW, Hejnol A, Matus DQ, Pang K, Browne WE, Smith SA, Seaver E, Rouse GW, Obst M, Edgecombe GD, et al. Broad phylogenomic sampling improves resolution of the animal tree of life. Nature. 2008;452:745–749. doi: 10.1038/nature06614. [DOI] [PubMed] [Google Scholar]
- 22.Hackett JD, Yoon HS, Li S, Reyes-Prieto A, Rümmele SE, Bhattacharya D. Phylogenomic analysis supports the monophyly of cryptophytes and haptophytes and the association of rhizaria with chromalveolates. Mol. Biol. Evol. 2007;24:1702–1713. doi: 10.1093/molbev/msm089. [DOI] [PubMed] [Google Scholar]
- 23.Hibbett DS, Binder M, Bischoff JF, Blackwell M, Cannon PF, Eriksson OE, Huhndorf S, James T, Kirk PM, Lücking R, et al. A higher-level phylogenetic classification of the Fungi. Mycol. Res. 2007;111:509–547. doi: 10.1016/j.mycres.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 24.Hsing M, Cherkasov A. Indel PDB: a database of structural insertions and deletions derived from sequence alignments of closely related proteins. BMC Bioinformatics. 2008;9:293. doi: 10.1186/1471-2105-9-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Z, Xing C, Wang L, Gong B, Liu H. IndelFR: a database of indels in protein structures and their flanking regions. Nucleic Acids Res. 2012;40:D512–D518. doi: 10.1093/nar/gkr1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen FC, Chen CJ, Chuang TJ. INDELSCAN: a web server for comparative identification of species-specific and non-species-specific insertion/deletion events. Nucleic Acid Res. 2007;35:W633–W638. doi: 10.1093/nar/gkm350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 28.Baldauf SL, Palmer JD. Animals and fungi are each other’s closest relatives: congruent evidence from multiple proteins. Proc. Natl Acad. Sci. USA. 1993;90:11558–11562. doi: 10.1073/pnas.90.24.11558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Belinky F, Cohen O, Huchon D. Large-scale parsimony analysis of metazoan indels in protein-coding genes. Mol. Biol. Evol. 2010;27:441–451. doi: 10.1093/molbev/msp263. [DOI] [PubMed] [Google Scholar]
- 30.Allard MW, Carpenter JM. On weighting and congruence. Cladistics. 1996;12:183–198. doi: 10.1111/j.1096-0031.1996.tb00008.x. [DOI] [PubMed] [Google Scholar]
- 31.Milinkovitch MC, LeDuc RG, Adachi J, Farnir F, Georges M, Hasegawa M. Effects of character weighting and species sampling on phylogeny reconstruction: a case study based on DNA sequence data in cetaceans. Genetics. 1996;144:1817–1833. doi: 10.1093/genetics/144.4.1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goloboff PA, Carpenter JM, Arias JS, Esquivel DRM. Weighting against homoplasy improves phylogenetic analysis of morphological data sets. Cladistics. 2008;24:1–16. [Google Scholar]
- 33.Young ND, Healy J. GapCoder automates the use of indel characters in phylogenetic analysis. BMC Bioinformatics. 2003;4:6. doi: 10.1186/1471-2105-4-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Redelings BD, Suchard MA. Incorporating indel information into phylogeny estimation for rapidly emerging pathogens. BMC Evol. Biol. 2007;7:40. doi: 10.1186/1471-2148-7-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Simmons MP, Müller K, Norton AP. The relative performance of indel-coding methods in simulations. Mol. Phylogenet. Evol. 2007;44:724–740. doi: 10.1016/j.ympev.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 36.Simmons MP, Ochoterena H. Gaps as characters in sequence-based phylogenetic analyses. Syst. Biol. 2000;49:369–381. [PubMed] [Google Scholar]
- 37.Müller K. SeqState: primer design and sequence statistics for phylogenetic DNA datasets. Appl. Bioinformatics. 2005;4:65–69. doi: 10.2165/00822942-200504010-00008. [DOI] [PubMed] [Google Scholar]