Abstract
Similarities in the 3D patterns of amino acid side chains can provide insights into their function despite the absence of any detectable sequence or fold similarities. Search for protein sites (SPRITE) and amino acid pattern search for substructures and motifs (ASSAM) are graph theoretical programs that can search for 3D amino side chain matches in protein structures, by representing the amino acid side chains as pseudo-atoms. The geometric relationship of the pseudo-atoms to each other as a pattern can be represented as a labeled graph where the pseudo-atoms are the graph's nodes while the edges are the inter-pseudo-atomic distances. Both programs require the input file to be in the PDB format. The objective of using SPRITE is to identify matches of side chains in a query structure to patterns with characterized function. In contrast, a 3D pattern of interest can be searched for existing occurrences in available PDB structures using ASSAM. Both programs are freely accessible without any login requirement. SPRITE is available at http://mfrlab.org/grafss/sprite/ while ASSAM can be accessed at http://mfrlab.org/grafss/assam/.
INTRODUCTION
In biological macromolecules, the 3-dimensional (3D) structure determines the functionality of the molecule. Therefore, it has long been recognized that similarities in structure can be a valuable guide to similarities in function, even if there is no detectable sequence similarity (1). For this reason, tools and services that are able to detect similarities in folding between different protein structures, such as DALI (2), have been available since the 1990s. However, it has also been clear for decades that similar constellations of amino acid residues in unrelated proteins can give rise to similar chemical activity, even where there is no fold similarity, sequence similarity or common evolutionary precursor. The classic example of this convergent evolution at the atomic level is the ‘catalytic triad’ of an aspartate, a histidine and a serine which was found to occur in both chymotrypsin and subtilisin (3). The result of this association in these unrelated enzymes is that the serine becomes very nucleophilic and is able to catalyze peptide bond cleavage. Such 3D amino acid constellations are therefore of interest because they may be involved in key functions that can include structure stabilization, binding and catalysis. The recognition of such similarities may hence be a valuable guide to function.
The ability to search for specific 3D arrangements of amino acids can be especially useful for structural biologists especially for identifying residues of interest in newly solved structures. This can be of value in assigning function to proteins of unknown function, or in identifying ligands that bind to similar sites in different proteins. One important use of such searches lies in the area of structural genomics where a large number of structures of proteins with unknown functions have been solved. Several existing services that allow for 3D motif searching include ProFunc/JESS (4), GIRAF (5), PINTS (6), SPASM (7), RIGOR (7), SuMo (8), RASMOT-3D PRO (9) and SA-Mot (10).
We have previously described a program, amino acid pattern search for substructures and motifs (ASSAM) that successfully uses a graph theoretical approach to search for and identify 3D motifs in protein structures (11,12). Here, we present a web service that deploys two graph theoretical computer programs. The first is a new program called search for protein sites (SPRITE), which allows the 3D structure of a protein to be searched against a database of curated sites, and the second is ASSAM itself, which accepts a 3D amino acid pattern as a query for searching against a database of protein structures.
PROGRAMS AND METHODS
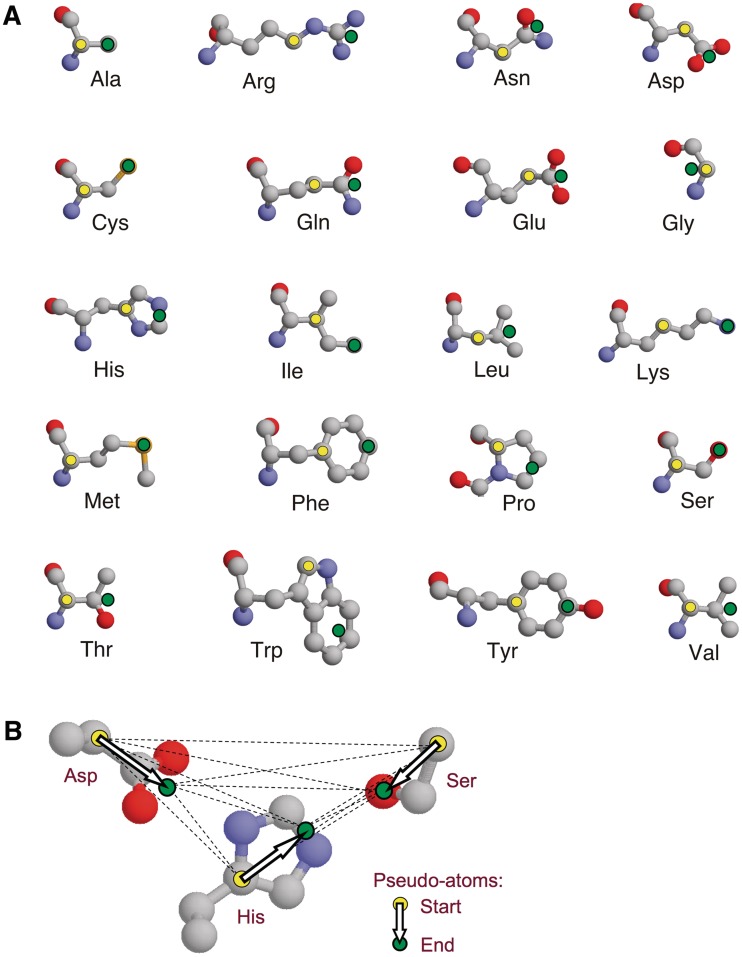
The basic concept behind the search methodology for both SPRITE and ASSAM has been described previously (11,12). Briefly, the protein structure is represented as a graph with the nodes representing individual amino acid side chains and the inter-node geometric relationships are the graphs. Each node consists of two pseudo-atoms which are used to generate a vector, and each such vector corresponds to one of the nodes in a graph (Figure 1A). The positions of the pseudo-atoms are chosen to emphasize the functional part of the side chain corresponding to that node. The geometric relationships between pairs of residues are defined in terms of distances calculated between the corresponding vectors, and these relationships correspond to the edges of a graph (Figure 1B). Specifically, if we let S, M and E denote the start, middle and end, respectively, of a vector, then the graph edges contain five parts, these being the SS, SE, ES, EE and MM distances (although only a subset of these five distances is normally used to specify a query pattern) (11).
Figure 1.
The side chain representation used in ASSAM and SPRITE. (A) The 20 amino acid types showing the locations of pseudo-atoms (yellow and green circles) used to represent side chains, with arrows representing the vectors between pseudo-atoms within a side chain. (B) Diagram of an aspartate–histidine–serine catalytic triad pattern showing with pseudo-atoms and vectors represented as in (A) and with dotted lines representing the distances between pseudo-atoms used in pattern matching. Diagram produced with Rasmol (13).
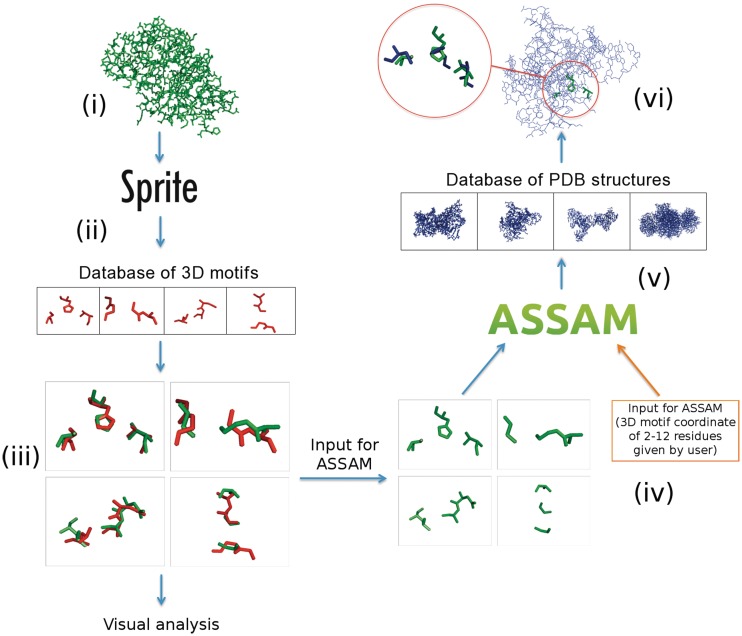
The current version of ASSAM, we report here, uses a maximal common subgraph (MCS) approach. This involves a fast initial screen using the Carraghan and Pardalos (1990) (14) clique detection algorithm to rapidly determine if any structural correspondences actually exist, followed, if appropriate, by the use of the Bron and Kerbosch (1973) (15) MCS algorithm to enumerate all the possible correspondences. SPRITE continues to use the Ullmann algorithm (16) of the original ASSAM but in a reversed approach in which a database of queries is compared with a single structure (Figure 2). The SPRITE and ASSAM programs provide separate outputs for both left-handed and right-handed superpositions, which, to our knowledge, is unique to these servers, and which can yield valuable chemical information (17) as discussed below.
Figure 2.
Diagram showing the input and output structure of SPRITE and ASSAM. (i) SPRITE accepts a whole structure in PDB format as input and (ii) compares it against a database of 3D motifs. (iii) The output is a list of amino acids in the query structure that matches to patterns in the database. (iv) ASSAM’s inputs can be either any 2–12 pattern residues given by the user, or selected from the hit residues in the query structure of a SPRITE search. (v) ASSAM then compares this pattern to representative structures in the PDB. (vi) The ASSAM output is a list of PDB structures that contain the query motif. An example is shown with the superposed hit residues identified and magnified as red circles.
The SPRITE program enables the user to examine a single complete protein structure in order to identify or annotate functional sites that have been documented in other structures. Such a utility can assist in providing insights into the potential function of proteins that yield no detectable sequence or fold similarity to existing examples in the databases and thus help direct function determination experiments. The ASSAM program, in contrast, enables the user to search the entire Protein Data Bank (18) for occurrences of a specific small amino acid motif. This can provide insights into the conservation and/or evolution of specific 3D arrangements such as catalytic sites. If required, the SPRITE and ASSAM programs can be used in series. As an example, a user can submit a query structure to identify which known motifs are present. Motifs of interest can immediately be submitted for an ASSAM search via the web interface to identify other structures where such motifs occur.
SPRITE: searching for sites in a protein structure query
The 3D SPRITE program accepts a PDB formatted file as input and this structure is searched against databases of sites annotated from X-ray crystallographic structures archived in the PDB. Perhaps the well-known example of such a database is the Catalytic Site Atlas (CSA) in which Porter et al. (19) used literature searches, hand annotation and homology searches to identify amino acids unequivocally involved in the catalytic activity of enzymes whose structures were stored in the PDB. In order to carry out a SPRITE search, the 3D arrangement of amino acids from the input file is converted into a graph representation that is then compared against the database of graph representations for patterns of sites in protein structures. Most SPRITE searches tested take <2 min to complete inclusive of upload times under the server's normal daily load.
Results are presented in a main menu (Figure 3) that provides a number of visualization options to the user, and hyperlinks to results of structural superpositions. When superposing two protein folds, there is a difference between, for example, a left-handed α-helical bundle and a right-handed one and they cannot be considered as equivalent. However, at the level of side chains, two non-evolutionarily related groupings of amino acids do not necessarily have to be of the same handedness in order to have the same chemical activity (17), they merely need to agree in terms of inter-residue distances. An example of this is the Asp–His–Ser catalytic triad in prolyl oligopeptidase, which is on the opposite hand to that in chymotrypsin (20), although it carries out the same peptidase function, albeit on a different substrate. Therefore, the program considers both right-handed and left-handed superpositions to be equally valid in principle and both are given in two separate results lists.
Figure 3.
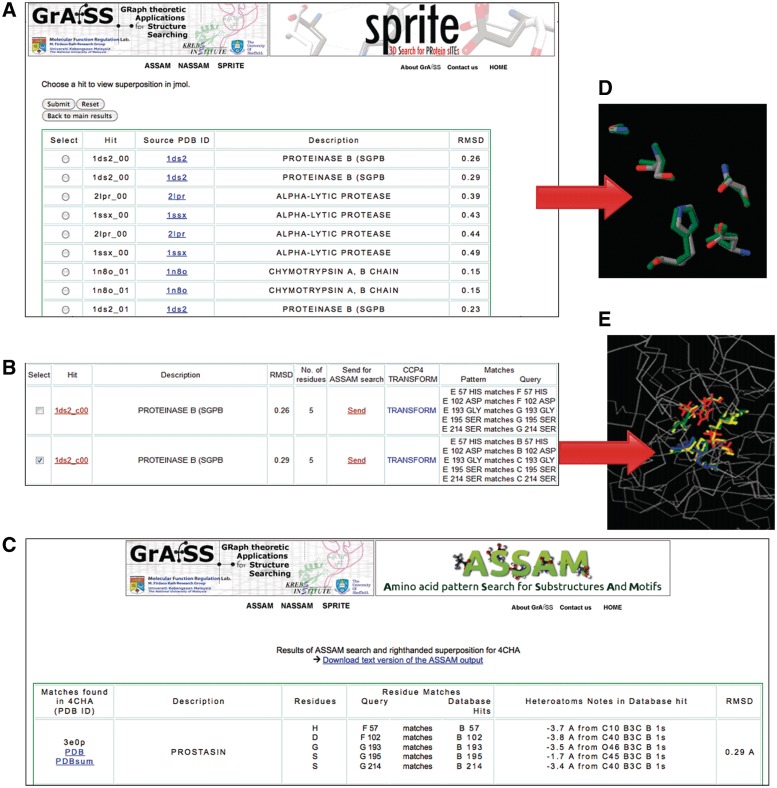
Snapshots of various pages from the web server showing the output of a SPRITE search using a serine proteinase structure (PDB ID: 4CHA). (A) List of motifs in the database that match to the query protein. (B) A detailed description of the results, showing the matched residues and the TRANSFORM command for CCP4; users can also opt to execute an ASSAM search on any hit of interest. (C) An example of an ASSAM search output, using the SPRITE output as a query pattern. (D) Jmol view of the superposition of a site of interest with the motif in the database that it matches to, which can be selected from page (A). (E) Jmol view showing the position of a site of interest relative to the query structure, which can be chosen from the page depicted in (B).
The three output visualization options for both the right- and left-handed superpositions are: (i) a list of the PDB structures which contain sites that match to sites in the query structure; (ii) a full list of matches that include RMSD values for the superpositions based on pseudo-atom positions, mapping of the query residues (number, chain and amino acid) to their database hits and a matrix for input as a TRANSFORM command in the CCP4 macromolecular crystallography software suite (http://www.ccp4.ac.uk; Figure 3B); and (iii) a list arranged by non-redundant matching sites in the query structure. The second option, which presents the full details of the hits, also allows the user to execute an ASSAM search using the residues in the query structure with hits to the SPRITE pattern database (Figure 2). All of the output browsing options listed also allow for visualization of the hits in a Jmol (http://www.jmol.org/) molecular viewer window. Users have the options of viewing superpositions of the query to the database matches (Figure 3D) or of viewing the residues in the query structure that have matches in the database (Figure 3E).
ASSAM: searching for a pattern in a structure database
The ASSAM program enables users to search for amino acid constellations of interest in a database of PDB structures (11). Users can input the coordinates of a 3D motif consisting of up to 12 amino acids as a PDB formatted file that is then used as a search query against a database of PDB structures. Depending on the server load and jobs already queued on the server, a typical ASSAM search for a three residue pattern took ∼6 min. The ASSAM results are presented as a list of hits for either right-handed or left-handed superpositions. Provided in the output is information regarding the residue matches to entries in the database, the RMSD of the matches, and information regarding the proximity of non-water hetero atoms to the pattern of interest, which may be a guide to the function of the residues detected (Figure 3C).
Databases associated with SPRITE and ASSAM
The primary source for patterns in the SPRITE search database are sites that have been annotated as catalytic sites in the CSA (19) (2667 patterns from the 20 January 2010 version). Additionally, the search database contains other 3D arrangements of amino acids that have been functionally characterized and curated, such as nucleotide binding sites (21) (382 patterns from the 26 March 2012 of 3D-Footprint), carbohydrate binding sites (22) (217 patterns from the November 2008 version of ProCarb) and patterns extracted from available literature. The versions of the data sets used are clearly presented for the user's reference. Users are able to view the list of entries currently available for searching by SPRITE. The ASSAM search database consists of the NCBI VAST non-redundant data set of (at present) 28 500 PDB structures, which is a list of sequence-dissimilar chains calculated on a P-value of 10 e−80. This excludes chains which are more than ca. 95% sequence identical to a better defined chain, although ASSAM searches the entire PDB deposition and not just the chain in question thus increasing the scope of the database. Users also have the option of executing an ASSAM search against a manually curated non-redundant version of the PDB consisting of 57 500 of the 80 400 available structures. For this data set, repeated structures such as mutants were manually removed, but versions of the same protein that do and do not contain ligands are retained. The SPRITE pattern database is periodically updated as new patterns become available while the ASSAM search database is updated monthly.
CASE STUDIES
The first example we present here is that of a hypothetical protein from Archaeoglobus fulgidus (PDB ID: 2OO2; UniProt: O28492). Sequence searches against the non-redundant sequence databases only identified hits to a number of putative uncharacterized proteins from various organisms. A DALI search (23) returned several matches, the most significant being an eight-heme nitrite reductase (PDB ID: 3F29) with a Z-value of 9.1. Several other nitrite reductases are also in the list of matches. A SPRITE search yielded two matches to a nitrite reductase (PDB ID: 1NID), with pseudo-atom RMSDs of 0.44 and 0.55 Å over two residues. Further analysis was done to gauge the significance of the hits, using CCRXP (24) and Metapocket 2.0 (25). CCRXP was designed to find clusters of conserved residues, which have been reported to play a crucial role in protein function (26), while Metapocket 2.0 is able to identify cavities on the protein surface that could indicate the location of an active site. The hit with the lower RMSD value, which is made up of the residues Phe43 and Gly47, occurs within a cluster of conserved residues consisting of six residues. This region overlaps with a cleft that potentially constitutes a ligand-binding site suggesting that 2OO2 might be a nitrite reductase or a similarly directed function.
The second example involves an intriguing ion-pair network from the structure of a Salmonella typhimurium sucrose specific porin ScrY (PDB ID: 1A0S) (27). In this motif, Arg 437 ion pairs with Glu 439 which ion pairs with Arg 441, which, in turn, also ion pairs with Glu 480, and then completes the square array of side chains by Glu 480 ions pairing back with Arg 437. This motif, which we called ‘RERE’, was submitted to an ASSAM search which yielded as expected the 1A0S structure in addition to other examples of sucrose specific porin (1A0T and 1OH2). Other non-sucrose-specific porin structures retrieved with good pseudo-atom superposition RMSD values include hits that also involve an Arg and a Glu from one subunit and a 2-fold related Arg and Glu from another subunit. From the results list, one interesting hit is for a bacteriophage RB69 DNA polymerase (PDB ID: 2ATQ, RMSD = 0.92 Å). We discuss this example further in comparison with other programs such as RASMOT-3D PRO and SPASM in the following section.
Comparisons with other methods
As mentioned previously above, other web servers exist (4–10) that allow analogous enquiries to those provided by SPRITE or ASSAM. Our comparative assessment revealed that the search methodologies and outputs presented by these programs differ in significant ways from the searches that SPRITE and ASSAM are able to carry out. In many respects, the currently available programs and SPRITE and ASSAM are complementary, and an integration of the results can provide useful insights or enable further investigations to be carried out.
The SPASM program (7) and RASMOT-3D PRO (9) service are similar to the ASSAM server in being able to search for 3D motifs in a database of structures. However, the SPASM and RASMOT-3D PRO structural representations are different compared to ASSAM. SPASM uses the Cα and the centre of gravity of the side chain while RASMOT-3D PRO uses the Cα and Cβ positions of each residue. In contrast, ASSAM and SPRITE use atoms from the functional part of the side chain itself (Figure 1) and are therefore more sensitive to the positions of the ends of side chains and less dependent on the main chain position. The use of the RERE motif from 1A0S, which we have described above, serves to demonstrate this effect. While the RASMOT-3D PRO was also able to retrieve the 1A0T structure, a sucrose-containing version of 1A0S, with an RMSD of 0.13, other matches in the results were not of the same RERE motif. For example, the next hits in the list provided by a RASMOT-3D PRO search were a GAAT motif (PDB ID: 1K42; RMSD = 0.28) and an ALER motif (PDB ID: 1X51; RMSD = 0.4).
The ASSAM search was able to retrieve a hit for a bacteriophage RB69 DNA polymerase (2ATQ) where the guanidium groups of two arginines (751 and 750) and the carboxylate groups of the two glutamates (747 and 745) overlap well with their 1A0S equivalents even though the positions of the main chain atoms are unrelated—a clear case of convergent evolution onto a similar side-chain motif from very different structural starting points (Figure 4). While programs, such as RASMOT-3D PRO and SPASM, may find other hits where there is a similar arrangement of Cα and Cβ vectors, ASSAM is able to identify patterns where the side chains are superposed even though the main chain positions are very different. This is therefore distinct and complementary to the other methods.
Figure 4.
(A) ASSAM-derived overlap of the RERE side chain pattern from DNA polymerase (PDB ID: 2ATQ, cyan side chain carbon atoms, white Cα and Cβ atoms) on the search pattern from S. typhimurium sucrose specific porin (PDB ID: 1A0S, green side chain carbon atoms, black Cα and Cβ atoms). The Cα atoms are shown as spheres to emphasize the dissimilarities in the main chain positions that nevertheless permit a similar constellation of side chains. (B) Details of the hit from the ASSAM web server output.
In addition, ASSAM, like SPRITE as mentioned earlier, separately retrieves both left- and right-handed overlaps with the search pattern. On the other hand, RASMOT-3D PRO is able to retrieve generic residue types, such as acidic for Glu or Asp or basic for Lys or Arg, and this is demonstrated in our earlier mention of the output retrieved by a RASMOT-3D PRO search using the RERE motif. We anticipate that the side-chain oriented pseudo-atom representation used in ASSAM will enable future generic residue type searches to be carried out. For example, the pseudo-atoms for an Asp are positioned on the CB and midpoint of OD1/OD2 whereas those for a Glu are on CG and OE1/OE2, where in both cases they correspond to the actual position of the carboxyl group. Once again this would be distinct from yet complementary to RASMOT-3D PRO.
The SA-Mot (10), MegaMotifBase (28) and ProFunc (4) servers are similar to the SPRITE web server because they allow a user to search a query structure for matches in a motif or pattern database. The SA-Mot server considers motifs that are part of loop formations and takes into consideration the sequence while a SPRITE search (like an ASSAM search) is independent of the sequence. In short, there are a diverse variety of different approaches to what is an important problem in structural biology.
SUMMARY
It can therefore be seen that in situations where a user is investigating the possible properties of a hypothetical protein structure with as yet uncharacterized function, a SPRITE search using that structure as a query is capable of identifying potential amino acid residues of functional importance. This information can facilitate the planning of experimental characterization strategies. Visual examination of a structure may also identify residues that imply a functional role. In such cases, ASSAM can be used to identify structures where similar patterns occur and can thus provide insights when they are repeated in structures with similar functions or properties.
FUNDING
Universiti Kebangsaan Malaysia [UKM-GGPM-KPB-101-2010 to M.F.-R.]; Ministry of Higher Education, Malaysia, National Science Fellowship (to N.N.). Funding for open access charge: University Kebangsaan Malaysia grant [UKM-DLP-2012-018].
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We gratefully acknowledge the Genome Computing Centre of the Malaysia Genome Institute for providing the computational infrastructure. We thank Mohd Noor Mat Isa and Hafiza Aida Ahmad for technical assistance with server operations.
REFERENCES
- 1.Artymiuk PJ, Poirrette AR, Rice DW, Willett P. A polymerase I palm in adenylyl cyclase? Nature. 1997;388:33–34. doi: 10.1038/40310. [DOI] [PubMed] [Google Scholar]
- 2.Holm L, Sander C. Dali: a network tool for protein structure comparison. Trends Biochem. Sci. 1995;20:478–480. doi: 10.1016/s0968-0004(00)89105-7. [DOI] [PubMed] [Google Scholar]
- 3.Warshel A, Naray-Szabo G, Sussman F, Hwang JK. How do serine proteases really work? Biochemistry. 1989;28:3629–3637. doi: 10.1021/bi00435a001. [DOI] [PubMed] [Google Scholar]
- 4.Laskowski RA, Watson JD, Thornton JM. ProFunc: a server for predicting protein function from 3D structure. Nucleic Acids Res. 2005;33:W89–W93. doi: 10.1093/nar/gki414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kinjo AR, Nakamura H. Comprehensive structural classification of ligand-binding motifs in proteins. Structure. 2009;17:234–246. doi: 10.1016/j.str.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 6.Stark A, Sunyaev S, Russell RB. A model for statistical significance of local similarities in structure. J. Mol. Biol. 2003;326:1307–1316. doi: 10.1016/s0022-2836(03)00045-7. [DOI] [PubMed] [Google Scholar]
- 7.Kleywegt GJ. Recognition of spatial motifs in protein structures. J. Mol. Biol. 1999;285:1887–1897. doi: 10.1006/jmbi.1998.2393. [DOI] [PubMed] [Google Scholar]
- 8.Jambon M, Andrieu O, Combet C, Deleage G, Delfaud F, Geourjon C. The SuMo server: 3D search for protein functional sites. Bioinformatics. 2005;21:3929–3930. doi: 10.1093/bioinformatics/bti645. [DOI] [PubMed] [Google Scholar]
- 9.Debret G, Martel A, Cuniasse P. RASMOT-3D PRO: a 3D motif search webserver. Nucleic Acids Res. 2009;37:W459–W464. doi: 10.1093/nar/gkp304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nuel G, Regad L, Martin J, Camproux AC. Exact distribution of a pattern in a set of random sequences generated by a Markov source: applications to biological data. Algorithms Mol. Biol. 2010;5:15. doi: 10.1186/1748-7188-5-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Spriggs RV, Artymiuk PJ, Willett P. Searching for patterns of amino acids in 3D protein structures. J. Chem. Inf. Comput. Sci. 2003;43:412–421. doi: 10.1021/ci0255984. [DOI] [PubMed] [Google Scholar]
- 12.Poirrette AR, Artymiuk PJ, Grindley HM, Rice DW, Willett P. Structural similarity between binding sites in influenza sialidase and isocitrate dehydrogenase: implications for an alternative approach to rational drug design. Protein Sci. 1994;3:1128–1130. doi: 10.1002/pro.5560030719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sayle RA, Milner-White EJ. RASMOL: biomolecular graphics for all. Trends Biochem. Sci. 1995;20:374. doi: 10.1016/s0968-0004(00)89080-5. [DOI] [PubMed] [Google Scholar]
- 14.Carraghan R, Pardalos PM. An exact algorithm for the maximum clique problem. Oper. Res. Lett. 1990;9:375–382. [Google Scholar]
- 15.Bron C, Kerbosch J. Algorithm 457: finding all cliques of an undirected graph. Commun. ACM. 1973;16:575–577. [Google Scholar]
- 16.Ullmann JR. An algorithm for subgraph isomorphism. J. ACM. 1976;23:31–42. [Google Scholar]
- 17.Garavito RM, Rossmann MG, Argos P, Eventoff W. Convergence of active center geometries. Biochemistry. 1977;16:5065–5071. doi: 10.1021/bi00642a019. [DOI] [PubMed] [Google Scholar]
- 18.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Porter CT, Bartlett GJ, Thornton JM. The Catalytic Site Atlas: a resource of catalytic sites and residues identified in enzymes using structural data. Nucleic Acids Res. 2004;32:D129–D133. doi: 10.1093/nar/gkh028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fulop V, Bocskei Z, Polgar L. Prolyl oligopeptidase: an unusual beta-propeller domain regulates proteolysis. Cell. 1998;94:161–170. doi: 10.1016/s0092-8674(00)81416-6. [DOI] [PubMed] [Google Scholar]
- 21.Contreras-Moreira B. 3D-footprint: a database for the structural analysis of protein-DNA complexes. Nucleic Acids Res. 2010;38:D91–D97. doi: 10.1093/nar/gkp781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Malik A, Firoz A, Jha V, Ahmad S. PROCARB: a database of known and modelled carbohydrate-binding protein structures with sequence-based prediction tools. Adv. Bioinformatics. 2010 doi: 10.1155/2010/436036. 436036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holm L, Rosenstrom P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ahmad S, Keskin O, Mizuguchi K, Sarai A, Nussinov R. CCRXP: exploring clusters of conserved residues in protein structures. Nucleic Acids Res. 2010;38:W398–W401. doi: 10.1093/nar/gkq360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Z, Li Y, Lin B, Schroeder M, Huang B. Identification of cavities on protein surface using multiple computational approaches for drug binding site prediction. Bioinformatics. 2011;27:2083–2088. doi: 10.1093/bioinformatics/btr331. [DOI] [PubMed] [Google Scholar]
- 26.DeLano WL. Unraveling hot spots in binding interfaces: progress and challenges. Curr. Opin. Struct. Biol. 2002;12:14–20. doi: 10.1016/s0959-440x(02)00283-x. [DOI] [PubMed] [Google Scholar]
- 27.Forst D, Welte W, Wacker T, Diederichs K. Structure of the sucrose-specific porin ScrY from Salmonella typhimurium and its complex with sucrose. Nat. Struct. Biol. 1998;5:37–46. doi: 10.1038/nsb0198-37. [DOI] [PubMed] [Google Scholar]
- 28.Pugalenthi G, Suganthan PN, Sowdhamini R, Chakrabarti S. MegaMotifBase: a database of structural motifs in protein families and superfamilies. Nucleic Acids Res. 2008;36:D218–D221. doi: 10.1093/nar/gkm794. [DOI] [PMC free article] [PubMed] [Google Scholar]