Abstract
Knowledge of all molecular interactions that potentially take place in the cell is a key for a detailed understanding of cellular processes. Currently available interaction data, such as protein–protein interaction maps, are known to contain false positives that inevitably diminish the accuracy of network-based inferences. Interaction confidence scoring is thus a crucial intermediate step after obtaining interaction data and before using it in an interaction network-based inference approach. It enables to weight individual interactions according to the likelihood that they actually take place in the cell, and can be used to filter out false positives. We describe a web tool called IntScore which calculates confidence scores for user-specified sets of interactions. IntScore provides six network topology- and annotation-based confidence scoring methods. It also enables the integration of scores calculated by the different methods into an aggregate score using machine learning approaches. IntScore is user-friendly and extensively documented. It is freely available at http://intscore.molgen.mpg.de.
INTRODUCTION
A detailed map of all interactions that take place between biomolecules in the cell promises unprecedented insight into biological processes in health and disease (1–4). Motivated by this, a number of techniques have been developed and applied to infer different types of interactions, such as physical protein–protein interactions (PPIs) (5,6) or genetic interactions (7,8). Numerous databases have been developed to store the resulting interaction data (9) and to assemble detailed interactome maps for human and for other species. However, current maps often contain considerable amounts of false positives resulting mainly from experimental or curation errors (10–12). Therefore, assigning confidence scores to interactions is a requirement for both data quality assessment of new data and predictive network analyses that use large interaction data sets. Several methods have been proposed that assess the confidence of individual interactions within a given interaction network (13,14). One group of methods use additional knowledge about the interactions or their participants like gene ontology (GO), pathway, gene expression or sequence homology information, while others exploit the topology of the network data as such for confidence assessment (14). Topology-based methods are the tools of choice when additional data are missing or biased (e.g. if the organism from which the interactions originate is not sufficiently studied). For most of the methods proposed in the literature, no publicly available software implementations are available. In addition, we have recently proposed a network topology-based confidence scoring method called cluster-based assessment of PPI confidence (CAPPIC; Kamburov et al., submitted). Notably, CAPPIC was shown to compare favorably to other topology-based methods on an extensive benchmark of several yeast PPI networks.
To provide the research community with a tool for interaction confidence scoring, we have developed IntScore (http://intscore.molgen.mpg.de). It enables the application of six different methods for interaction confidence scoring including CAPPIC, two topology-based methods proposed by other authors (15,16), as well as three annotation-based scoring schemes (all methods are described below). Furthermore, IntScore supports the integration of confidence scores calculated by different methods into an aggregate score through different supervised machine learning approaches. Such integration is useful because different methods tend to exploit different topological and functional features of interactions, and thus, an aggregate score is expectedly more reliable. The output of IntScore is a weighted variant of the input network where the interaction scores calculated by each user-selected method are provided. The output further includes score distribution histograms and a correlation table that gives a clue about the pairwise coherency of scoring methods when applied to the given network. Confidence scores calculated by IntScore can be used, e.g. as interaction weights when the network is utilized in computational methods that handle probabilistic input data. Otherwise, confidence scores can be used as a filtering criterion to remove interactions that are likely to be false positive. The input, output and the scoring methods provided by IntScore are extensively documented on the website.
A comparable tool for interaction confidence scoring is PSISCOREweb (17). It serves as a common gateway for web services that match a list of user-specified interactions against public databases such as IntAct (18) and MINT (19) and assign pre-defined scores from those databases to the interactions found. Similarly, the recently published tool HIPPIE (20) assigns pre-calculated scores based on experimental evidence to those interactions from a user-specified network that have been published previously. PRINCESS (21) is another complementary interaction confidence evaluation system with multiple data sources, comprising interaction homology, domain interactions, gene ontology (GO) annotation, genome context, co-expression and network motifs in public interaction databases. Like PSISCOREweb and HIPPIE, PRINCESS also critically depends on the extensiveness and quality of gene annotations for calculating confidence scores. Accordingly, scores cannot be calculated for interactions involving non-annotated genes, and reliability of the scores depends on gene annotation accuracy. In contrast, IntScore calculates confidence scores dynamically rather than assigning pre-calculated scores: For example, all topology-based methods (available in our tool and in none of the others mentioned above) use exclusively the structure of the user-specified network to assess the confidence of its interactions regardless of whether the interactions are present in public databases or not. Thus, IntScore is not restricted to organisms for which e.g. accurate interactome maps are publicly available. As for the annotation-based methods, IntScore allows a greater flexibility than previous tools regarding e.g. the input accession number namespace (IntScore supports 11 identifier types while HIPPIE and PRINCESS support only two types each), the method-specific parameter settings (e.g. the GO semantic similarity measure is selectable by the user) and the interaction types (IntScore supports both physical and genetic interactions, while the other tools are focused on physical interactions only). Similar to PSISCOREweb and PRINCESS, IntScore also offers GO semantic similarity measures (detailed below); however, in addition to the calculated confidence scores, IntScore optionally exports the respective GO annotations. Likewise, IntScore supports a literature evidence method similar to PSISCOREweb, but IntScore optionally also exports the according PubMed identifiers. Reliability of the literature evidence and the pathway co-occurrence methods provided by our tool is achieved through utilizing ConsensusPathDB, a fairly comprehensive interaction meta-database developed by us (22). Another advantage of IntScore over PSISCOREweb and HIPPIE is the possibility to automatically calculate aggregate scores. Although PRINCESS does also calculate an aggregate score, the integration method and the positive/negative reference sets are fixed while in IntScore they can be specified by the user. This flexibility is very important, considering that the scientific community is currently debating on generic gold standard interaction sets (11,23,24).
CONFIDENCE SCORING METHODS
IntScore provides overall six interaction confidence scoring methods. Three of them exploit exclusively the topology of the given input interaction network for confidence assessment, while three are based on annotation of interacting genes/proteins and their interactions. The provided methods are described below.
Topology-based methods
CAPPIC
The CAPPIC method proposed by us exploits the network’s inherent modular structure for assessing the confidence of individual interactions. It assigns interaction scores according to the graphical co-clustering of interactions. Intuitively, low confidence is assigned to interactions that disagree with modularity of biological networks and high confidence to those that comply with it. CAPPIC determines algorithmic parameters intrinsically and does not require any parameter input or reference sets for confidence scoring. Details on the method will be described elsewhere (Kamburov et al., submitted).
Common neighbors
In this approach proposed by Goldberg and Roth (15), interaction confidence is calculated as the level of enrichment of common network neighbors of interacting proteins. It is quantified by the hypergeometric test P-value given the number of common neighbors and total network neighbors of both interacting proteins. The rationale behind the approach is based on the existence of densely connected local neighborhoods (neighborhood cohesiveness property) in biological networks (25,26). Real PPIs are expected to meet the network cohesiveness property more frequently than false positives.
Geometric embedding
In this method proposed by Kuchaiev et al. (16), interaction networks are embedded into a low-dimensional Euclidean space based on network metrics (shortest path length) and then confidence values are calculated depending on the Euclidean distance between proteins within that space.
Annotation-based methods
Literature evidence
For every user-specified pair of genes/proteins, IntScore examines whether and how often (i.e. in how many different publications) the corresponding interaction is reported in the literature. The number of different publications reporting an interaction has often been used as a reliable measure of interaction confidence (11,27). The interaction meta-database ConsensusPathDB (22) is used as the source of literature annotation. ConsensusPathDB currently integrates 30 public interaction and pathway resources and thus represents a fairly comprehensive annotation basis. We have shown previously that ConsensusPathDB can be used for evidence mining of user-specified interactions (28).
GO semantic similarity
This method calculates the similarity of the GO (29) terms annotated to interacting genes/proteins, taking into account the overall structure of the GO to assess the specificity of shared annotations (30). The rationale behind this method is that genes/proteins that interact in the cell are more likely to participate in the same biological process and cellular compartment (31). Accordingly, true positive interactions are expected to have more similar GO annotations and get higher scores than false positives.
Pathway co-occurrence
This method checks pairs of genes/proteins for co-occurrence in biochemical pathways. As in the GO semantic similarity method, the rationale is that interacting genes/proteins in the cell are more likely to be involved in the same biological process. The key difference to the GO semantic similarity measure is that the pathway annotation basis is ConsensusPathDB (instead of GO), which currently contains 3281 curated pathways from 12 public pathway databases. Also, the pathway co-occurrence results in a binary indicator whereas the GO semantic similarity measure provides a continuous score.
Integration of confidence scores from different methods
IntScore offers the possibility to combine confidence scores calculated by different methods into an aggregate score. This is generally useful because the different scoring methods tend to capture different topological or functional features of interactions (an exception is that the GO biological process semantic similarity score and the pathway co-occurrence evidence are somewhat redundant to each other). Thus, an aggregate confidence score should be more reliable. To enable score integration, two disjoint subsets from the input interactions must be distinguished by the user as positive and negative reference sets, respectively. They are used by IntScore to train either a support vector machine or a logistic regression model (selectable by the user), where the confidence scores calculated by the separate methods are treated as interaction features. The model is then used to predict confidence values for the whole input network. Although integral for quality assessment of interaction data, the generation of suitable gold standard positive and negative interaction sets is still a daunting task (11,23,24). Due to the different and complementary nature of PPI inference techniques, there are no generic positive and negative reference sets that are suitable for data quality assessment. Rather, the different nature of interaction data can most reliably be assessed with reference sets accounting for the data properties, such as whether binary interaction data, co-complex data or genetic interaction data need to be assessed (32). Due to lack of a generic, optimal solution, the choice for reference sets is up to the user. However, if no reference sets are at hand, IntScore can be used to generate such. A strategy to do this suggested here is to apply the literature evidence and GO semantic similarity methods on the network to distinguish interactions that are likely to be true positives (e.g. the ones reported in three or more publications) and gene/protein pairs that are likely to be non-interacting (e.g. randomized gene/protein pairs without literature evidence, where the two proteins are located in different compartments as per GO cellular component annotation) (13,23). These sets of interactions can then be used as positive and negative interaction sets for a subsequent IntScore request. In this case, it should be noted that the scoring methods used to generate the positive and negative interaction sets must not be included in a subsequent score integration because this would bias the aggregate score.
INPUT, OUTPUT AND SCALING BEHAVIOR
Input
The input for IntScore is a binary interaction network, i.e. a list of interactions each having strictly two participants (genes/proteins). It can either be uploaded as a flat file or pasted in the respective input field (Figure 1A). If any annotation-based methods are to be used, the interaction participants should be consistently annotated with Entrez, Ensembl, RefSeq, UniProt, HGNC, CYGD, SGD, MGI accession numbers or official gene symbols. After providing an input network, the user should select one or more methods for confidence scoring. The description of each parameter for each method can be found on the IntScore web site and its documentation. If multiple methods are selected, their output can be integrated into an aggregate score. In this case, the user should additionally provide two disjoint subsets from the input interaction set that represent positive and negative (i.e. non-interacting), interaction sets, accordingly. As discussed above, IntScore can be used also to generate such sets.
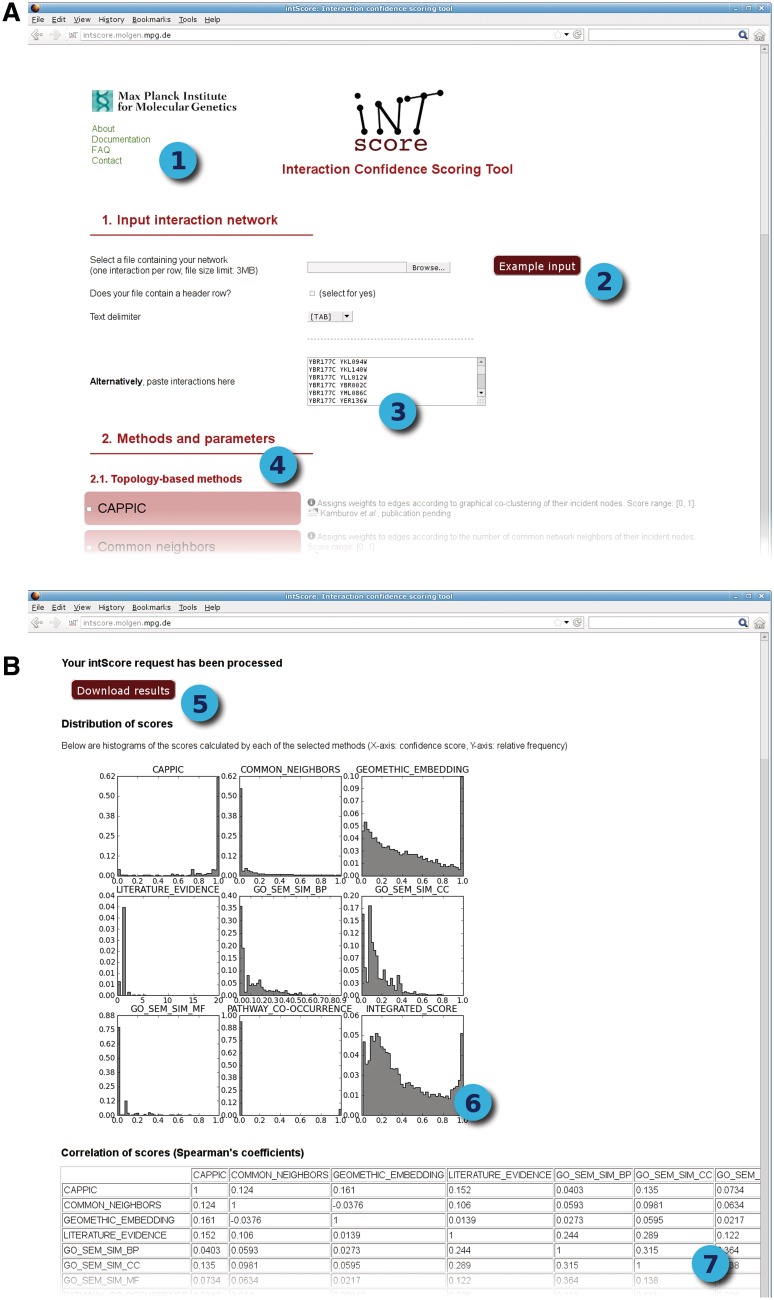
Figure 1.
Screenshots of the input form (A) and results page (B) of IntScore. 1) Links to documentation and general information about IntScore; 2) Button for loading example data (high-quality interaction network from (34) with 3% randomly rewired interactions); 3) Input field where interactions can be pasted; 4) List of methods provided by IntScore, selectable by the user; 5) Download button for results of IntScore; 6) Histograms showing the score distribution of each user-selected method; 7) Correlation table showing the Spearman correlation coefficients for scores calculated by different methods.
Output
IntScore returns a zip archive file containing (i) the confidence scores calculated by the user-selected methods for each interaction as a tab-delimited file (‘network.scored.tsv’); (ii) the distributions of scores calculated by each method visualized with an image (‘scoreDistributions.png’); (iii) a table in tab-delimited format that contains the Spearman correlation coefficients between scores resulting from different methods, if more than one method is selected (‘correlationTable.tsv’). Depending on the user's request, the zip file may also contain (iv) the PubMed identifiers of publications that report each interaction (‘network.pubmedIds.tsv’); (v) the GO terms annotated to interactors (e.g. ‘network.goBPannotation.tsv’ for biological process annotations); (vi) the names and database sources of biochemical pathways where the pair of interacting genes/proteins are found together (‘network.containingPathways.tsv’); and (vii) a plot resulting from the clustering granularity (inflation parameter) estimation procedure of CAPPIC, if the respective option of CAPPIC is selected (‘cappic_inflationScanPlot.png’) [relevant only for expert users interested in more details about CAPPIC, cf. (Kamburov et al., submitted)]. The scores calculated by the different methods have different ranges, which are specified in the short description of each method on the IntScore input form (Figure 1A). However, scores obtained with each of the six methods increase with increasing interaction confidence. Aggregate scores correspond to probability estimates, i.e. a low aggregate score indicates that the corresponding interaction is likely false positive (or interactors lack biological information, if annotation-based methods are integrated), whereas a high aggregate score indicates that an interaction is likely to be true. Apart from providing the zip file described above, the web server displays the distributions of scores, a score correlation table, method-specific details (e.g. the training accuracy of the score integration procedure, if selected), as well as a process log (Figure 1B). The weighted network output by IntScore can be used directly as input for computational analysis tools such as Cytoscape (33).
Scaling behavior
The runtime of IntScore depends primarily on (i) the size of the provided network, (ii) which scoring methods are selected by the user, as well as (iii) the parameter settings of the selected methods. The runtime performance of topology-based methods additionally depends on the intrinsic structure of the network, e.g. CAPPIC may perform differently for networks with the same size but with different modularity. Other factors influencing the runtime of IntScore are how busy the processing servers are at the time of the submission, and the user’s internet connection. Systematic runtime performance estimation is complicated by the high number of available methods and parameters. Instead, we summarized in Supplementary Figure S1, the runtimes for 10 example networks with different sizes within the range of a typical interaction screen (although IntScore can handle networks containing >105 interactions), in order to give users an intuition about how long it will take to process their request. Notably, IntScore features an optional e-mail notification system that sends results to the user as soon as they are ready.
EXAMPLE APPLICATION
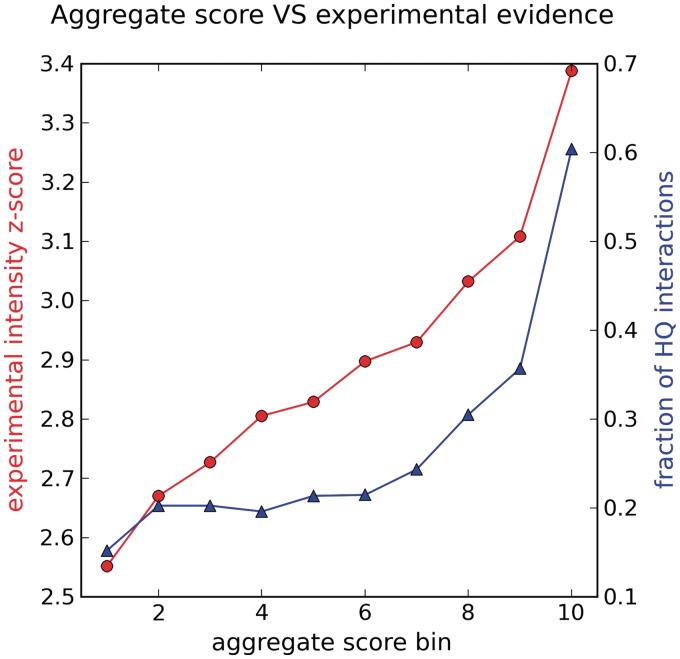
We applied IntScore on a proteome-scale yeast network derived with the protein-fragment complementation assay (PCA) (34). The original network consisted of 10 230 interactions among 2293 proteins. For each interaction, the authors have provided a z-score corresponding to the significance of the PCA intensity readout. Furthermore, they have distinguished a set of 2770 high-quality interactions among 1124 proteins based on experimental interaction evidence. We used the largest connected component of the complete network excluding self-interactions as input for IntScore. This network consisted of 9605 interactions among 2238 proteins (Supplementary Data set 1). Our goal was to assess the consistency between an aggregate score calculated by IntScore that integrates topology-based and annotation-based methods with the independent, experimental evidence provided by the authors of the study. To integrate the scores calculated by the separate methods in IntScore, we first created positive and negative reference sets. As a positive interaction set, we distinguished 391 interactions from the input network that had more than two literature references. This was done using the literature evidence method provided by IntScore. To construct a negative set (i.e. non-interacting protein pairs), we randomly rewired 3% of the input network, preserving the nodes’ degrees (35). In a subsequent IntScore request, all confidence methods were executed on the input network using standard parameters. To avoid bias, the literature evidence method was excluded from the calculation of the aggregate score, because this method had been used to construct the positive reference set. The distributions of the scores calculated by the separate methods and the aggregate score are depicted in Figure 1B. We binned all interactions ordered by aggregate score into 10 equally sized bins and calculated the mean experimental intensity z-score and the fraction of high-quality interactions (as per experimental evidence) in each bin. Figure 2 shows that the aggregate interaction confidence score based on topological and annotation features correlated well with the independent experimental interaction evidence. The Spearman correlation coefficient between the aggregate score and the experimental z-score was 0.31 (P < 10−6). The fraction of high-quality interactions also increased with increasing aggregate score. For example, 12 out of the 100 interactions with the lowest aggregate score and 85 out of the 100 interactions with the highest aggregate score have been denoted high-quality as per experimental evidence. This example application demonstrates that an aggregate score combining topological and annotation features of interactions corroborates experimental interaction evidence.
Figure 2.
Experimental interaction score (red line, left-hand side y-axis) and fraction of high-quality interactions distinguished as per experimental evidence (blue line, right-hand side y-axis) are plotted against interaction bins with increasing aggregate score as calculated by IntScore (x-axis). The input network corresponds to an in vivo PCA map of yeast (34). HQ: high quality.
CONCLUSION
IntScore is a web server for confidence scoring of biological interactions. It provides six methods for confidence scoring, as well as the possibility to integrate the method-specific scores. IntScore can serve experimentalists to increase the quality of data produced by interaction screens and assess the performance of those screens, and can help computational biologists to increase the reliability of network-based inferences by controlling the accuracy of the input interaction data.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figure 1 and Supplementary Data set 1.
FUNDING
The European Commission under its Seventh Framework Programme with the grant diXa [283775]; German Ministry of Education and Research [MedSys PREDICT, 0315428A; NGFNp, NeuroNet-TP3, 01GS08171]; Max Planck Society. Funding for open access charge: European Commission.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
IntScore was developed exclusively with open-source software. Contributors are gratefully acknowledged.
REFERENCES
- 1.Ideker T, Sharan R. Protein networks in disease. Genome Res. 2008;18:644–652. doi: 10.1101/gr.071852.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barabasi A-L, Gulbahce N, Loscalzo J. Network medicine: a network-based approach to human disease. Nat. Rev. Genet. 2011;12:56–68. doi: 10.1038/nrg2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vidal M, Cusick ME, Barabasi A-L. Interactome networks and human disease. Cell. 2011;144:986–998. doi: 10.1016/j.cell.2011.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ideker T, Krogan NJ. Differential network biology. Mol. Syst. Biol. 2012;8:565. doi: 10.1038/msb.2011.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stelzl U, Wanker EE. The value of high quality protein-protein interaction networks for systems biology. Curr. Opin. Chem. Biol. 2006;10:551–558. doi: 10.1016/j.cbpa.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 6.Gstaiger M, Aebersold R. Applying mass spectrometry-based proteomics to genetics, genomics and network biology. Nat. Rev. Genet. 2009;10:617–627. doi: 10.1038/nrg2633. [DOI] [PubMed] [Google Scholar]
- 7.Beyer A, Bandyopadhyay S, Ideker T. Integrating physical and genetic maps: from genomes to interaction networks. Nat. Rev. Genet. 2007;8:699–710. doi: 10.1038/nrg2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boone C, Bussey H, Andrews BJ. Exploring genetic interactions and networks with yeast. Nat. Rev. Genet. 2007;8:437–449. doi: 10.1038/nrg2085. [DOI] [PubMed] [Google Scholar]
- 9.Bader GD, Cary MP, Sander C. Pathguide: a pathway resource list. Nucleic Acids Res. 2006;34:D504–D506. doi: 10.1093/nar/gkj126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Levy ED, Landry CR, Michnick SW. How perfect can protein interactomes be? Sci. Signal. 2009;2:pe11. doi: 10.1126/scisignal.260pe11. [DOI] [PubMed] [Google Scholar]
- 11.Venkatesan K, Rual J-F, Vazquez A, Stelzl U, Lemmens I, Hirozane-Kishikawa T, Hao T, Zenkner M, Xin X, Goh K-I, et al. An empirical framework for binary interactome mapping. Nat. Methods. 2009;6:83–90. doi: 10.1038/nmeth.1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cusick ME, Yu H, Smolyar A, Venkatesan K, Carvunis A-R, Simonis N, Rual J-F, Borick H, Braun P, Dreze M, et al. Literature-curated protein interaction datasets. Nat. Methods. 2009;6:39–46. doi: 10.1038/nmeth.1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Suthram S, Shlomi T, Ruppin E, Sharan R, Ideker T. A direct comparison of protein interaction confidence assignment schemes. BMC Bioinformatics. 2006;7:360. doi: 10.1186/1471-2105-7-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chua HN, Wong L. Increasing the reliability of protein interactomes. Drug Discov. Today. 2008;13:652–658. doi: 10.1016/j.drudis.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 15.Goldberg DS, Roth FP. Assessing experimentally derived interactions in a small world. Proc. Natl Acad. Sci. USA. 2003;100:4372–4376. doi: 10.1073/pnas.0735871100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuchaiev O, Rasajski M, Higham DJ, Przulj N. Geometric de-noising of protein-protein interaction networks. PLoS Comput. Biol. 2009;5:e1000454. doi: 10.1371/journal.pcbi.1000454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aranda B, Blankenburg H, Kerrien S, Brinkman FSL, Ceol A, Chautard E, Dana JM, De Las Rivas J, Dumousseau M, Galeota E, et al. PSICQUIC and PSISCORE: accessing and scoring molecular interactions. Nat. Methods. 2011;8:528–529. doi: 10.1038/nmeth.1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kerrien S, Aranda B, Breuza L, Bridge A, Broackes-Carter F, Chen C, Duesbury M, Dumousseau M, Feuermann M, Hinz U, et al. The IntAct molecular interaction database in 2012. Nucleic Acids Res. 2012;40:D841–D846. doi: 10.1093/nar/gkr1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Licata L, Briganti L, Peluso D, Perfetto L, Iannuccelli M, Galeota E, Sacco F, Palma A, Nardozza AP, Santonico E, et al. MINT, the molecular interaction database: 2012 update. Nucleic Acids Res. 2012;40:D857–D861. doi: 10.1093/nar/gkr930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schaefer MH, Fontaine J-F, Vinayagam A, Porras P, Wanker EE, Andrade-Navarro MA. HIPPIE: integrating protein interaction networks with experiment based quality scores. PLoS One. 2012;7:e31826. doi: 10.1371/journal.pone.0031826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li D, Liu W, Liu Z, Wang J, Liu Q, Zhu Y, He F. PRINCESS, a protein interaction confidence evaluation system with multiple data sources. Mol. Cell Proteom. 2008;7:1043–1052. doi: 10.1074/mcp.M700287-MCP200. [DOI] [PubMed] [Google Scholar]
- 22.Kamburov A, Pentchev K, Galicka H, Wierling C, Lehrach H, Herwig R. ConsensusPathDB: toward a more complete picture of cell biology. Nucleic Acids Res. 2011;39:D712–D717. doi: 10.1093/nar/gkq1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jansen R, Gerstein M. Analyzing protein function on a genomic scale: the importance of gold-standard positives and negatives for network prediction. Curr. Opin. Microbiol. 2004;7:535–545. doi: 10.1016/j.mib.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 24.Chatr-Aryamontri A, Ceol A, Licata L, Cesareni G. Protein interactions: integration leads to belief. Trends Biochem. Sci. 2008;33:241–242. doi: 10.1016/j.tibs.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 25.Strogatz SH. Exploring complex networks. Nature. 2001;410:268–276. doi: 10.1038/35065725. [DOI] [PubMed] [Google Scholar]
- 26.Watts DJ, Strogatz SH. Collective dynamics of small-world networks. Nature. 1998;393:440–442. doi: 10.1038/30918. [DOI] [PubMed] [Google Scholar]
- 27.von Mering C, Krause R, Snel B, Cornell M, Oliver SG, Fields S, Bork P. Comparative assessment of large-scale data sets of protein-protein interactions. Nature. 2002;417:399–403. doi: 10.1038/nature750. [DOI] [PubMed] [Google Scholar]
- 28.Pentchev K, Ono K, Herwig R, Ideker T, Kamburov A. Evidence mining and novelty assessment of protein-protein interactions with the ConsensusPathDB plugin for Cytoscape. Bioinformatics. 2010;26:2796–2797. doi: 10.1093/bioinformatics/btq522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu G, Li F, Qin Y, Bo X, Wu Y, Wang S. GOSemSim: an R package for measuring semantic similarity among GO terms and gene products. Bioinformatics. 2010;26:976–978. doi: 10.1093/bioinformatics/btq064. [DOI] [PubMed] [Google Scholar]
- 31.Oliver S. Guilt-by-association goes global. Nature. 2000;403:601–603. doi: 10.1038/35001165. [DOI] [PubMed] [Google Scholar]
- 32.Yu H, Braun P, Yildirim MA, Lemmens I, Venkatesan K, Sahalie J, Hirozane-Kishikawa T, Gebreab F, Li N, Simonis N, et al. High-quality binary protein interaction map of the yeast interactome network. Science. 2008;322:104–110. doi: 10.1126/science.1158684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smoot ME, Ono K, Ruscheinski J, Wang P-L, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27:431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tarassov K, Messier V, Landry CR, Radinovic S, Serna Molina MM, Shames I, Malitskaya Y, Vogel J, Bussey H, Michnick SW. An in vivo map of the yeast protein interactome. Science. 2008;320:1465–1470. doi: 10.1126/science.1153878. [DOI] [PubMed] [Google Scholar]
- 35.Maslov S, Sneppen K, Zaliznyak A. Detection of topological patterns in complex networks: correlation profile of the internet. Phys. A Stat. Mech. Appl. 2004;333:529–540. [Google Scholar]