Abstract
With recent improvements in DNA sequencing and sample extraction techniques, the quantity and quality of metagenomic data are now growing exponentially. This abundance of richly annotated metagenomic data and bacterial census information has spawned a new branch of microbiology called comparative metagenomics. Comparative metagenomics involves the comparison of bacterial populations between different environmental samples, different culture conditions or different microbial hosts. However, in order to do comparative metagenomics, one typically requires a sophisticated knowledge of multivariate statistics and/or advanced software programming skills. To make comparative metagenomics more accessible to microbiologists, we have developed a freely accessible, easy-to-use web server for comparative metagenomic analysis called METAGENassist. Users can upload their bacterial census data from a wide variety of common formats, using either amplified 16S rRNA data or shotgun metagenomic data. Metadata concerning environmental, culture, or host conditions can also be uploaded. During the data upload process, METAGENassist also performs an automated taxonomic-to-phenotypic mapping. Phenotypic information covering nearly 20 functional categories such as GC content, genome size, oxygen requirements, energy sources and preferred temperature range is automatically generated from the taxonomic input data. Using this phenotypically enriched data, users can then perform a variety of multivariate and univariate data analyses including fold change analysis, t-tests, PCA, PLS-DA, clustering and classification. To facilitate data processing, users are guided through a step-by-step analysis workflow using a variety of menus, information hyperlinks and check boxes. METAGENassist also generates colorful, publication quality tables and graphs that can be downloaded and used directly in the preparation of scientific papers. METAGENassist is available at http://www.metagenassist.ca.
INTRODUCTION
Comparative metagenomics is a newly emerging branch of metagenomics that involves the comparison of complex microbial populations across different samples using either shotgun metagenomic sequence data or amplified 16S rRNA data. Comparisons of large metagenomic data sets only became possible with the development of lower cost, higher throughput Next-Gen DNA sequencing technologies (1). Even though comparative metagenomics is still in its infancy, its impact is starting to be felt in many areas of health and environmental science. For example, comparative metagenomics was recently used to determine that there are only three major ‘enterotypes’ associated with human gut microflora (2). Similar comparative metagenomics studies have identified distinct gut microflora differences between ‘lean’ and obese individuals (3). On the environmental science side, extensive comparative metagenomic studies of ocean-dwelling microbes have been used to help understand the survival strategies of different groups of prokaryotic picoplankton (4). These kinds of results point to the tremendous potential that comparative metagonomic studies could offer in helping to understand disease, to learn more about host–environment interactions, to enhance environmental remediation and to rationalize observations on ecological diversity. While the potential applications for comparative metagenomics seem to be almost endless, there is still a bottleneck with regard to how comparative metagenomic data are processed. Unlike other fields such as comparative genomics, comparative proteomics, comparative transcriptomics or comparative metabolomics, comparative metagenomics has barely entered the world of web-based servers. Indeed most of today’s comparative metagenomics tools either offer a limited selection of statistical tools or are command-line programs, sometimes requiring complex program installations or the use of separate programs for data visualization. Given that most microbiologists and many metagenomics researchers are not necessarily trained with such computer skills, there is a clear need to develop tools that are more accessible or user-friendly. Here we describe METAGENassist, a web server that provides a broad range of statistical tools for comparative metagenomics through an easy-to-use graphical interface. Not only is METAGENassist unique for being the first web server dedicated to comprehensive comparative metagenomics, it is also unique in its ability to perform automated phenotypic enrichment and in its ability to support sophisticated data analyses with this phenotypic information.
METAGENassist maintains a strict focus on supporting statistical comparisons across metagenomic samples. As result, METAGENassist leaves the task of sequence processing and taxonomic assignment to other well-established metagenomics programs, such as mothur (5), QIIME (6), MG-RAST (7), MEGAN (8) or other taxonomic assignment tools. In other words, METAGENassist is designed for use after the raw sequence data has been processed using any of the above programs. Once the raw data have been processed, users can submit their taxonomic abundance data (obtained from either 16S rRNA or shotgun metagenomic experiments) in a generic comma-separated value format (.csv) or in any of several common formats produced from the programs mentioned above, including the STAMP comparative metagenomics package (9). In addition to the taxonomic abundance data, a metadata file (containing sample metadata such as collection conditions, depth, temperature, pH, salinity, tissue, pathology or patient information) is also required. This permits the facile mapping of sample conditions and sample attributes onto METAGENassist’s statistical plots.
METAGENassist is designed for researchers with at least some basic statistical knowledge. Through a user-friendly and intuitive web interface, it leads users through a guided workflow or analysis pipeline with tips, check-boxes, FAQs and pointers to ensure that users complete the right steps in the right order. After being guided through some preliminary data processing and data normalization steps, users are then directed to select from a range of univariate and multivariate statistical analyses, including fold change analysis, t-tests, ANOVA, principal components analysis (PCA), partial least squares discriminant analysis (PLS-DA) and hierarchical clustering. Each of these analyses is explained in some detail through tutorials and FAQ pages. In addition to these standard statistical offerings, METAGENassist also supports a number of unique or advanced machine learning and statistical methods for supervised learning and feature selection, such as random forest and support vector machine (SVM) methods. These methods have seldom been used in metagenomic analysis, but they have proven to be very useful in related ‘omics’ fields such as transcriptomics (10,11) and metabolomics (12).
METAGENassist also stands apart from existing comparative metagenomics packages through its extensive use of automated taxonomic-to-phenotypic mapping. This mapping is done through a unique microbial phenotype database developed specifically for METAGENassist. The database contains phenotypic information for more than 11 000 microbial species, including nearly 1800 fully sequenced microbes (13,14). The phenotype information covers nearly 20 phenotypic categories for each microbe, including oxygen requirements, preferred temperature range, metabolism, energy source(s), habitat, GC content (for sequenced microbes), genome size (for sequenced microbes) and other properties. This information is maintained using a categorical, controlled vocabulary to facilitate multivariate data processing. It is also updated regularly through information drawn from BACMAP (14), GOLD (15) and other NCBI microbial taxonomy resources (16). This phenotypic enrichment allows researchers to use METAGENassist to examine their data from a number of novel perspectives. Instead of being limited to analyzing differences between samples in terms of taxonomic ‘distance’, users can compare microbial samples on the basis of their oxygen requirements, their preferred habitat, their GC content or other features. One other metagenomics program (MEGAN) does provide limited (eight categories) taxonomic-to-phenotypic mapping for the creation of bar charts and pie charts. However, METAGENassist goes somewhat further by allowing the phenotypic information to be directly integrated into PCA, PLS-DA or other clustering/classification plots. This phenotypic enrichment step effectively leverages the existing and accumulated scientific knowledge about known bacterial phenotypes in a way that could not easily be done by most microbiologists and certainly not done by any existing metagenomics programs.
PROGRAM DESCRIPTION
METAGENassist is designed to be a user-friendly, full-featured comparative metagenomics server that can be used by a broad range of researchers, from bench biologists with a basic understanding of statistics to bioinformaticians with more advanced experience. In particular, it uses clickable buttons, simple pull-down menus, fillable text boxes and a navigational tree to guide users through various univariate and multivariate analyses in a logical, step-by-step manner. It is perhaps useful to provide a short synopsis of these steps before describing them in more detail (below). All METAGENassist analyses begin with an initial data upload step. After uploading a suitably formatted taxonomic profile file and a corresponding metadata file, users can use the data integrity check module to detect errors or inconsistencies. The same module can also be used to impute missing values for any input data. After the data-checking step is complete, users are presented with several options for data filtering and data normalization. Once these data preprocessing steps are finished, it is then possible to move directly to the data analysis modules. Users can easily navigate between different types of analysis options by clicking the nodes of the navigational tree on the left-hand panel. This navigational tree lists all the available categories of univariate analysis, multivariate analysis, cluster analysis, classification and feature selection that can be performed on the input data. Upon completion of these analyses, users can download their results and any figures generated from their work using the result download page. A more detailed description of these five steps is given below.
Step 1: Data upload
After clicking on the ‘Click here to start’ link on the METAGENassist homepage, users are taken immediately to the data upload page. METAGENassist requires users to submit both a taxonomic profile file (containing taxonomic abundance information for their samples) and a file containing metadata information for each sample. Various options are given for submitting taxonomic profile information. In particular, the server can read files generated from mother (5), QIIME (6), MG-RAST (7), MEGAN (8) or STAMP (9). Alternately, users may choose to upload their taxonomic data in a generic comma-separated value (.csv) format. All taxonomic assignments should be made using other programs before submission to METAGENassist. It is also assumed that data in the taxonomic abundance file are ‘correct’ in that the samples have been sequenced to appropriate levels of coverage or depth and that appropriate rarefaction tests have been performed.
Detailed instructions are given on the data formats page on how to use the supported programs to generate an appropriate file(s) for submission. Since sequence reads are often matched with varying degrees of confidence to different taxonomic ranks (species being the lowest rank, kingdom/domain being the highest rank), submitted taxonomic profiles can include a mix of taxonomic assignments to different ranks. Valid taxonomic names can be either a list of taxonomic ranks going down the taxonomic hierarchy or simply the lowest matched rank. For the metadata file (i.e. the file containing sample collection conditions, depth, temperature, pH, salinity, tissue, pathology or patient information, etc.), a simple comma-separated value (.csv) format is supported. Metadata should consist of qualitative categorical labels rather than quantitative data, although both are allowed. Users can categorize their quantitative metadata in their prepared file as they see fit, preferably with a minimum of three samples for each distinct sample label to be included in the statistical calculations. More detailed information about these issues is given on the METAGENassist data formats page.
Step 2: Taxonomic-to-phenotypic mapping
METAGENassist uses a phenotype database containing phenotypic information for 11 000 or more bacterial and archaebacterial species listed in the NCBI microbial taxonomy. The phenotypic information includes oxygen requirements, temperature range, biotic relationship, habitat, host type, pathogenicity, energy source, metabolism, disease association, gram stain, cell shape, cell arrangement, sporulation, chromosome shape, plasmid shape, number of membranes, flagella, motility, GC content (for sequenced organisms) and genome size (for sequenced organisms). The phenotype database is largely built from the annotations found in BacMap (12,13), an up-to-date electronic atlas of over 1700 annotated prokaryotic genomes and expanded or extrapolated to include other species through extensive text mining and intelligent inference by taxonomic relationships. PubMed (16), GOLD (15) and other resources were also mined to provide information for metabolism and disease associations.
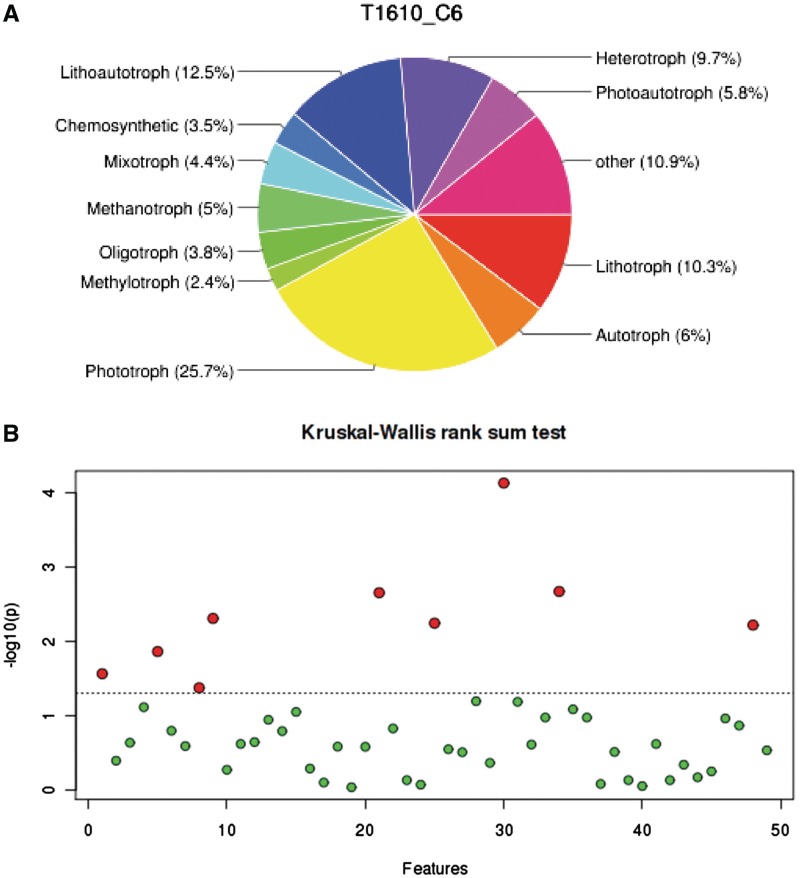
Using the names for species, genus or other classes provided in the user-submitted taxonomic profile data, METAGENassist performs name normalization and taxonomic rank identification using the standard NCBI microbial taxonomy. When non-taxonomic names are encountered (e.g. code numbers or ‘unclassified’), a valid taxonomic name at a higher rank level can be used if this information is given in a multi-level taxonomic identifier (see data upload section above). Using METAGENassist’s taxonomic-to-phenotype mapping utility allows phenotypes to be mapped directly to microbial species. For input taxonomic names that are only specified above the species rank (representing an unidentified species but a known genus or a known family), phenotype information is used whenever it can be safely inferred up the taxonomic tree based on the phenotypes of known members of a given taxonomic group. By combining the mapped phenotype information with the taxonomic abundance data in the submitted taxonomic profile, new microbial census tables are automatically created for each of the nearly 20 phenotype categories. These sample-by-phenotype tables form the basis for all downstream phenotype-oriented plots and statistical analyses in METAGENassist. Figure 1A shows a pie-chart summary of the annotated phenotype information derived for a typical metagenomic sample.
Figure 1.
Sample output from METAGENassist for phenotype mapping and univariate analysis. (A) A pie chart showing the breakdown of energy source phenotypes for a single sample after taxonomic mapping; (B) One-way ANOVA plot with red dots indicating significant features scoring above a given threshold (the horizontal dotted line).
Step 3: Data normalization
At this stage, the uploaded data are compiled into different tables with samples in rows and features in columns. To proceed further with any statistical analysis, it is necessary to ‘normalize’ the data. Normalization refers to a process of transforming or scaling the data to follow a Gaussian (or Normal, bell-shaped) distribution. Two types of data normalization procedures are available—row-wise normalization and column-wise normalization. The former is used to normalize or scale each sample so the samples are more comparable to each other. This is particularly important when there are systemic differences within samples (i.e. biomass, sequencing depth, etc., that are unrelated to conditions of interest). Several commonly used normalization or scaling methods are available in METAGENassist, including normalization to the median or total sum, normalization to a reference sample and normalization against a reference feature. Column-wise normalization aims to make each feature more comparable in magnitude to the other, thereby yielding a more Normal/Gaussian distribution. This can be done via log transformation, auto-scaling, Pareto scaling and range scaling. Row- and column-wise normalization procedures are often applied sequentially to make the data values more compatible with downstream statistical analysis. A diagnostic plot is provided so that users can check how well the data normalization procedures are working in terms of generating a Normal or Gaussian distribution for the input data.
Step 4: Statistical data analysis
This module offers a collection of well-established statistical and machine learning algorithms that have proven to be very useful in high-dimensional (i.e. omic) data analysis. These include the following.
Univariate analysis
Univariate analyses are widely used to obtain a ranked list of potentially important features (i.e. taxa, oxygen requirements and genome size/complexity) that have changed significantly due to the different conditions under study. Univariate methods are simple and easy to understand. Consequently, they are usually the first to be applied before trying to use more sophisticated approaches. Several widely used methods are offered in METAGENassist, including fold change analysis, t-tests, Mann–Whitney tests, ANOVA, Kruskal–Wallis tests, volcano plots as well as correlation analysis. Figure 1B shows a sample output from an ANOVA analysis. Users can click the ‘view selected features’ link above each graphical summary to get more detailed information about the features that have been selected.
Multivariate analysis
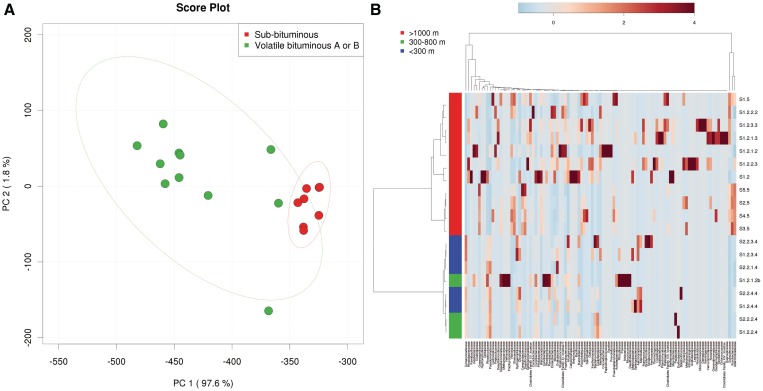
Since most metagenomic data sets are high dimensional, we implemented two widely used approaches for dimension reduction and visualization—PCA and PLS-DA. PCA is an unsupervised clustering method for transforming a complex collection of data points such that the important properties of the sample can be more simply displayed on a two-dimensional cluster/scatter plot. On a PCA plot, the first, and most significant vector or discriminating feature set is called the first principal component (X-axis) and the second most significant vector is called the second principal component (Y-axis). Two clusters on a PCA plot indicate that there are some significant differences between the two sets of samples. PLS-DA is a supervised classification method designed to enhance the separation between the groups by rotating the PCA components to achieve maximum separation. It improves the identification and understanding of the variables that most effectively distinguish the groups being studied. A false conclusion regarding separation achieved between the two groups can sometimes occur in PLS-DA. To minimize the possibility that the observed separation is due to chance, permutation testing is commonly used. In permutation testing, random re-labeling of the data is performed and then the PLS-DA is re-run again. This is repeated (2000 times), with different random labelings. In both PLS-DA and PCA, the values of these components are called scores, while the loadings are the coefficients corresponding to the projection directions. METAGENassist provides a number of graphical summary techniques commonly used for PCA and PLS-DA. Users can specify which axes should be plotted to observe the patterns between different components. Both 2D and 3D views are implemented. As an unsupervised method, PCA offers a more descriptive approach for identifying or visualizing patterns or clusters. In particular, data points can be colored according to their phenotypic labels. Figure 2A shows a PCA score plot based on metabolism phenotype labels from a sample data set. The most important features contributing to the separation can be easily identified from the corresponding loading plot. METAGENassist supports two commonly used PLS-DA feature importance measures—variable importance in projection (VIP) and the weighted sum of PLS-regression coefficients. The most significant features are presented as a dot chart, with each feature ranked by its significance measure along with the corresponding abundance profiles plotted side by side to facilitate interpretation.
Figure 2.
Sample outputs from METAGENassist for multivariate analysis. (A) A PCA plot of environmental coal samples for samples versus metabolism phenotypes, with colored groupings according to sample-associated coal type. The dotted ellipses indicate the 95% confidence intervals. (B) A heatmap combined with agglomerative hierarchical clustering using the same coal sample data.
Clustering
Cluster analysis allows researchers to identify samples that are more similar to each other based on a defined distance measure derived from their feature abundance profiles. There are two main approaches to clustering—hierarchical clustering and partitional clustering. Agglomerative hierarchical clustering starts by treating each individual as a separate cluster and then proceeds to combine them until all samples belong to a single cluster. METAGENassist’s hierarchical clustering implementation allows users to select among four different distance measures (Jensen-Shannon divergence, Euclidean distance, Pearson’s correlation, and Spearman’s rank correlation) and four clustering methods (average linkage, complete linkage, single linkage and Ward’s linkage) to perform the analysis. The result is presented as a dendrogram in combination with a heatmap. Users need to decide on a cutoff level to place different samples into different groups. In contrast to hierarchical clustering, partitional clustering directly decomposes the data set into specific numbers of clusters. METAGENassist offers two widely used methods—k-means clustering and self-organizing maps (SOM)—both of which allow users to experiment with different cluster numbers and visualize the results. k-means clustering attempts to create k clusters in which each sample belongs to the cluster with the nearest mean. SOM, also known as Kohonen mapping, is a type of artificial neural network that uses an iterative process to map nonlinear statistical dependencies between high-dimensional data into simple geometric relationships on a two-dimensional grid. The clusters from both k-means and SOM are presented by plotting all samples in each cluster as line graphs overlapping each other to facilitate visual comparison of their aggregated abundance profiles.
Supervised classification
Supervised methods are becoming increasingly important for discrimination analysis in comparative metagenomics (17). There are usually two main goals—the identification of important features (i.e. taxa or mapped phenotypes) and the prediction or assignment of attribute labels for new samples. METAGENassist offers two supervised classification methods that have proven to be robust for high-dimensional ‘omics’ data analysis—random forest (RF) (18) and SVM (19). RF uses an ensemble of decision trees, each of which is created by training on a bootstrap sample. During tree construction, about one-third of the instances are left out and used as test samples to calculate the classification error [also known as out-of-bag (OOB) error]. During the process, RF also generates a very useful feature importance measure known as ‘the mean decrease in accuracy’, which corresponds to the increase of the OOB error when it is permuted. Prediction is based on the majority vote of the all the classification trees. In addition to these functions, METAGENassist’s RF analysis of also provides two other useful functions: (i) a data overview based on multidimensional scaling (MDS) and (ii) outlier detection (20). In contrast to the RF algorithm, SVM classification aims to separate two classes of data by means of a maximum margin hyperplane. In order to achieve this, the SVM algorithm finds a nonlinear decision function in the input space by mapping the data into a higher dimensional feature space. METAGENassist’s SVM analysis was implemented based on an approach called recursive SVM feature selection and sample classification (R-SVM) as described by Zhang et al. (21). R-SVM uses SVM for both classification and for selecting a subset of relevant features according to their relative contribution in the classification, using cross-validation error rates. The least important features are eliminated in the subsequent steps. This process is done recursively to create a series of SVM models. The features used by the best model are considered to be important and are ranked by their frequencies of being selected in the model. Please note that this approach currently only supports binary classification analysis using a linear kernel.
Step 5: Results download
When users finish their analyses and click the download link, the processed data and all associated plots or images (in PNG format) are made available for downloading. A zip file is created containing all the data generated during the different analyses. Users are encouraged to download all the results immediately after they finish their analysis. All files are automatically deleted from the server after 72 h.
IMPLEMENTATION
METAGENassist was implemented using the same general framework as MetaboAnalyst (12). Briefly, the web interface was developed using Java Server Faces (http://java.sun.com/javaee/javaserverfaces, JSF) technology. The backend statistical computing and visualization operations were mainly carried out using functions from the R and Bioconductor packages. The integration between Java and R was established through the Rserve package (http://www.rforge.net/Rserve). The microbial phenotype database and the taxonomic mapping utility was based on resources compiled from BacMap (13,14), GOLD (15) and NCBI (16). The pie-chart visualization tool was implemented using Google Chart Tools. In addition, Perl scripts were used to perform the taxonomy-to-phenotype mapping, while Python scripts were used for parsing the input from different formats.
METAGENassist is currently hosted on GlassFish (version 3) installed on a Linux operating system (Debian GNU/Linux 6.0.4). The server is equipped with Intel Core 2 Quad Q9550 2.83 GHz CPU and 8GB of physical memory. The web application is platform independent and has been tested successfully under Linux, Windows and Mac operating systems. R (version 2.15.0) is currently installed on the same machine with latest Bioconductor release 2.10.
COMPARISON TO OTHER AVAILABLE TOOLS
Several stand-alone programs (but only one other web server) have been developed that support limited comparative metagenomic functions using either 16S rRNA surveys or shotgun metagenomic datasets (see Table 1). For 16S analysis, the stand-alone program known as mothur (5) supports several kinds of multivariate statistical analyses, such as principal coordinates analysis (PCoA) and Nonmetric Multidimensional Scaling (NMDS). Mothur can also perform a univariate test known as analysis of molecular variance (AMOVA) and homogeneity of molecular varriance (HOMOVA). Mothur has recently added the Metastats tool (22) for determining which taxa are differentially abundant between two groups of samples. Like mothur, QIIME (6) allows users to create PCoA and NMDS plots. QIIME is a stand-alone program that also generates Heatmap plots, performs Jackknife tests and supports the random forest supervised learning technique. While both mothur and QIIME are excellent stand-alone programs for the analysis of 16S rRNA samples, they lack a convenient or user-friendly graphical user interface. Both require command-line inputs and the pipeline nature of QIIME presents extra challenges for software installation.
Table 1.
Comparison of comparative metagenomic software
| METAGENassist | Mothur | QIIME | MG-RAST | MEGAN 4 | STAMP 2.0 | |
|---|---|---|---|---|---|---|
| Platform | Web server | Command-line | Command-line | Web server | Desktop and command-line | Desktopa and command-line |
| Graphical user interface | Yes | No | Nob | Yes | Yes | Yes |
| Compare 16S rRNA samples | Yes | Yes | Yes | Yes | Yes | Yesc |
| Taxonomy-centered analysis of shotgun data | Yes | No | Partial (using imported file) | Yes | Yes | Yes |
| Sequence annotation | No | Yes | Yes | Yes | Using input BLAST file | No |
| Univariate statistics | Fold change analysis, t-tests, volcano plots, ANOVA, Kruskal–Wallis H-test, correlation analysis | Metastats(22), AMOVA, HOMOVA | Paired t-test, Pearson correlation, G-test, ANOVA | No | Directed Homogeneity test (23) | t-tests, ANOVA, Kruskal–Wallis H-test |
| Multivariate statistics | PCA, PLS-DA | PCoA, PCA, NMDS | PCoA, NMDS | PCoA | No | PCA |
| Clustering | Dendrograms, heatmaps, K-means, SOM | Dendrograms, heatmaps | Heatmaps | Heatmap- dendrogram | No | No |
| Supervised classification | Random forest, SVM | No | Random forest | No | No | No |
| Interactive normalization | Yes | No | No | No | No | No |
| Metadata overlay | Yes | No | Yes | Yes | No | Yes |
| Taxonomy-to-phenotype mapping | Yes | No | No | No | Yes | No |
aAs of the writing of this paper, use on Mac and Linux required either virtualization or compilation from source code.
bSome graphical interaction is available on script-generated HTML pages.
cDoes not read files from mothur or QIIME.
For taxonomy-centered comparisons across shotgun metagenomic samples, the programs MG-RAST (7) and MEGAN 4 (8) offer excellent data visualization options through nicely designed graphical user interfaces. However, both have a somewhat limited complement of statistical tests. In particular, MG-RAST (which is a web server) supports only PCoA plots, while MEGAN (which is a stand-alone program) only allows users to visualize how significantly taxonomic abundances differ between exactly two samples or groups of samples, using a specialized ‘Directed Homogeneity test’ (23). STAMP (9) is another stand-alone program that offers a convenient graphical interface for a variety of statistical tests including multi-group, two-group and two-sample comparisons, focusing on PCA and univariate methods. More detailed program comparisons are provided in Table 1.
CONCLUSION
There is an increasing need for tools for comparative metagenomics that are statistically robust, easily accessible and easy to use—especially for those without specialized training in computer programming or a comprehensive statistical knowledge. METAGENassist was developed in response to these needs. In particular, METAGENassist provides users with a broad range of statistical tools through an easy-to-use graphical interface that is accessible over the web. By designing METAGENassist so that it readily accepts data files generated from some of the most popular metagenomics tools and by coupling taxonomic-to-phenotypic mapping with both standard and newly emerging statistical analyses, we believe we have created a system that will make comparative metagenomics far more accessible, far more powerful and far more informative for members of the metagenomics community.
FUNDING
Genome Alberta, Genome Canada, the Canadian Institutes of Health Research (CIHR) and Alberta Innovates. Funding for open access charge: Genome Alberta.
Conflict of interest statement. None declared.
REFERENCES
- 1.Metzker ML. Sequencing technologies—the next generation. Nat. Rev. Genet. 2010;11:31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- 2.Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, Fernandes GR, Tap J, Bruls T, Batto JM, et al. Enterotypes of the human gut microbiome. Nature. 2011;473:174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 4.Yooseph S, Nealson KH, Rusch DB, McCrow JP, Dupont CL, Kim M, Johnson J, Montgomery R, Ferriera S, Beeson K, et al. Genomic and functional adaptation in surface ocean planktonic prokaryotes. Nature. 2010;468:60–66. doi: 10.1038/nature09530. [DOI] [PubMed] [Google Scholar]
- 5.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meyer F, Paarmann D, D'Souza M, Olson R, Glass EM, Kubal M, Paczian T, Rodriguez A, Stevens R, Wilke A, et al. The metagenomics RAST server—a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics. 2008;9:386. doi: 10.1186/1471-2105-9-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huson DH, Mitra S, Ruscheweyh HJ, Weber N, Schuster SC. Integrative analysis of environmental sequences using MEGAN4. Genome Res. 2011;21:1552–1560. doi: 10.1101/gr.120618.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Parks DH, Beiko RG. Identifying biologically relevant differences between metagenomic communities. Bioinformatics. 2010;26:715–721. doi: 10.1093/bioinformatics/btq041. [DOI] [PubMed] [Google Scholar]
- 10.Herrero J, Al-Shahrour F, Diaz-Uriarte R, Mateos A, Vaquerizas JM, Santoyo J, Dopazo J. GEPAS: a web-based resource for microarray gene expression data analysis. Nucleic Acids Res. 2003;31:3461–3467. doi: 10.1093/nar/gkg591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reich M, Liefeld T, Gould J, Lerner J, Tamayo P, Mesirov JP. GenePattern 2.0. Nat. Genet. 2006;38:500–501. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- 12.Xia J, Psychogios N, Young N, Wishart DS. MetaboAnalyst: a web server for metabolomic data analysis and interpretation. Nucleic Acids Res. 2009;37:W652–W660. doi: 10.1093/nar/gkp356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stothard P, Van Domselaar G, Shrivastava S, Guo A, O'Neill B, Cruz J, Ellison M, Wishart DS. BacMap: an interactive picture atlas of annotated bacterial genomes. Nucleic Acids Res. 2005;33:D317–D320. doi: 10.1093/nar/gki075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cruz J, Liu Y, Liang Y, Zhou Y, Wilson M, Dennis JJ, Stothard P, Van Domselaar G, Wishart DS. BacMap: an up-to-date electronic atlas of annotated bacterial genomes. Nucleic Acids Res. 2012;40:D599–D604. doi: 10.1093/nar/gkr1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pagani I, Liolios K, Jansson J, Chen IM, Smirnova T, Nosrat B, Markowitz VM, Kyrpides NC. The Genomes OnLine Database (GOLD) v.4: status of genomic and metagenomic projects and their associated metadata. Nucleic Acids Res. 2012;40:D571–D579. doi: 10.1093/nar/gkr1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sayers EW, Barrett T, Benson DA, Bolton E, Bryant SH, Canese K, Chetvernin V, Church DM, Dicuccio M, Federhen S, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2012;40:D13–D25. doi: 10.1093/nar/gkr1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Knights D, Costello EK, Knight R. Supervised classification of human microbiota. FEMS Microbiol. Rev. 2011;35:343–359. doi: 10.1111/j.1574-6976.2010.00251.x. [DOI] [PubMed] [Google Scholar]
- 18.Breiman L. Random forests. Machine Learn. 2001;45:5–32. [Google Scholar]
- 19.Noble WS. What is a support vector machine? Nat. Biotechnol. 2006;24:1565–1567. doi: 10.1038/nbt1206-1565. [DOI] [PubMed] [Google Scholar]
- 20.Liaw A, Wiener M. Classification and regression by random forest. R. News. 2002;2:18–22. [Google Scholar]
- 21.Zhang X, Lu X, Shi Q, Xu XQ, Leung HC, Harris LN, Iglehart JD, Miron A, Liu JS, Wong WH. Recursive SVM feature selection and sample classification for mass-spectrometry and microarray data. BMC Bioinformatics. 2006;7:197. doi: 10.1186/1471-2105-7-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.White JR, Nagarajan N, Pop M. Statistical methods for detecting differentially abundant features in clinical metagenomic samples. PLoS Comput. Biol. 2009;5:e1000352. doi: 10.1371/journal.pcbi.1000352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mitra S, Klar B, Huson DH. Visual and statistical comparison of metagenomes. Bioinformatics. 2009;25:1849–1855. doi: 10.1093/bioinformatics/btp341. [DOI] [PubMed] [Google Scholar]