Abstract
The accuracy of atomistic biomolecular modeling and simulation studies depend on the accuracy of the input structures. Preparing these structures for an atomistic modeling task, such as molecular dynamics (MD) simulation, can involve the use of a variety of different tools for: correcting errors, adding missing atoms, filling valences with hydrogens, predicting pK values for titratable amino acids, assigning predefined partial charges and radii to all atoms, and generating force field parameter/topology files for MD. Identifying, installing and effectively using the appropriate tools for each of these tasks can be difficult for novice and time-consuming for experienced users. H++ (http://biophysics.cs.vt.edu/) is a free open-source web server that automates the above key steps in the preparation of biomolecular structures for molecular modeling and simulations. H++ also performs extensive error and consistency checking, providing error/warning messages together with the suggested corrections. In addition to numerous minor improvements, the latest version of H++ includes several new capabilities and options: fix erroneous (flipped) side chain conformations for HIS, GLN and ASN, include a ligand in the input structure, process nucleic acid structures and generate a solvent box with specified number of common ions for explicit solvent MD.
INTRODUCTION
Molecular modeling and simulations are routinely used to study the structure, function and activity of biomolecules (1–6). The accuracy of such modeling and simulations depends critically on the accuracy of the input structure (7). Approximately 88% of the structures in the protein data bank (PDB) (8) are determined by X-ray crystallography, which can not, in general, resolve positions of most hydrogen atoms. While the positions for most of the missing hydrogen atoms can be easily estimated using simple chemical rules, predicting the protonation states of titratable groups such as the side chains of ASP, GLU, ARG, LYS, TYR, HIS or CYS is non-trivial. Their protonation states depend on complex electrostatic interactions between these groups themselves and with the surrounding environment.
Many theoretical methods have been developed for predicting protonation states of titratable groups (9–23). While these methods vary in the details of the underlying physical models, they share one common feature—computational and algorithmic complexity. Hence, the computational process usually involves multiple non-trivial steps. There is often an additional complication arising from errors and inconsistencies in the input PDB structures, requiring significant ‘pre-processing’ of structures. As a result, predicting protonation equilibria (pK), adding missing protons to PDB structures and generating the necessary input files for molecular dynamics (MD) simulation can involve the use of multiple tools from different packages. The complexity of this computational ‘pipe-line’ makes it difficult for novice and even moderately experienced users to identify, install, learn and effectively use the appropriate tools, increasing the likelihood of errors and creating unnecessary barriers for the adoption of these tools. Even for experts, the careful setup required for MD simulations takes a considerable amount of time.
H++ (http://biophysics.cs.vt.edu/H++) is a freely available open-source web server for automating the addition of missing protons to the PDB structures based on predicted pK s of titratable groups, and generating basic parameter/topology and coordinate files for MD simulations (currently only in AMBER format). The calculations are based on the standard continuum solvent methodology (11), within the framework of the Poisson Boltzmann (PB) model (21,24). The server is intended for both experts and non-experts. Typical default values are used for processing parameters and options, although a limited subset of parameters and options can be modified on the input screen.
The original implementation of H++ was described in Gordon et al. (25). The present work summarizes the key features of H++ and describes in additional detail significant new enhancements since the original implementation, see the ‘Materials and Methods’ section below. The accuracy of the current version of H++ is presented in the ‘Results’ section.
MATERIALS AND METHODS
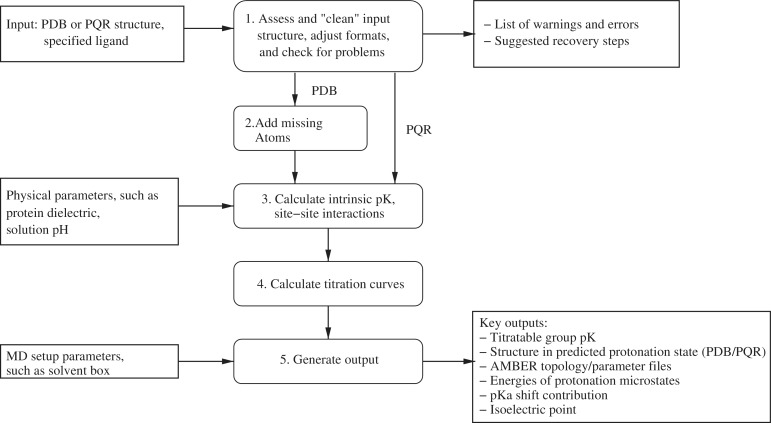
The input structure to be processed by H++ can be either in the PDB format, or alternatively in the PQR format, which is similar to the PDB format with atomic charges and radii included. Figure 1 shows the five main processing steps in H++, which are described below.
Figure 1.
Flowchart of computations performed by the H++ server described here.
Step 1: pre-process the input structure
Input PDB files can contain numerous errors and format inconsistencies, such as missing heavy atoms, suboptimal residue conformations and non-standard atom names. H++ attempts to make automatic, albeit conservative corrections for many of these problems when possible. Otherwise the errors are identified for possible manual correction. For example, the N and O atoms in the amide groups of ASN and GLN, and the N and C atoms in the imidazole ring of HIS cannot be easily distinguished from electron density maps. Thus, the assignment of these atoms in the PDB file may be (optionally) ‘flipped’ using the reduce algorithm that is based on an analysis of van der Waals contacts and H-bonding (26). An example of errors that are identified for manual correction are missing residues in the middle of protein chains. Input PDB files may also contain HETATM entries for solvent and ligand molecules; H++ removes these entries. Solvent molecules are removed by default because they are treated implicitly by the continuum solvent methodology used. Non-protein ligands are removed by default, but an option is now available to manually include many ligands and specific buried water molecules for processing, as described on the H++ site. Inclusion of buried waters has been shown to improve the accuracy of computed pK of nearby groups (27). For peptide, protein, DNA and RNA ligands, current AMBER force field parameters are used to add H atoms and assign atomic partial charges. For other organic ligands, H++ uses OpenBabel (28) to add H atoms, and atomic partial charges are assigned using the antechamber module from AmberTools (29) and the generalized AMBER force field (GAFF) parameter set. PDB structures may also contain residues with partial occupancy representing multiple possible conformations. Without manual intervention from the user, H++ selects the ‘A’ conformation and ignores all others.
An input PQR file, on the other hand, is assumed to have already been validated (e.g. in order to compute the atomic charges and radii included in the PQR file). Therefore, most error and consistency checks are bypassed for input PQR files. In addition, H++ requires AMBER compatible atom and residue names in the input PQR file.
Step 2: add missing atoms
In this step, missing heavy atoms and protons are added (assuming standard protonation states of all titratable groups at this stage), and atomic partial charges and radii [Bondi (30)] are assigned using the LEAP modules in AmberTools. The position of added atoms, and assignment of atomic partial charges and radii are based on the latest AMBER force field parameters (ff10). This step is bypassed for PQR files, (Figure 1) assuming that missing atoms have been added, and other consistency checks have been performed by the user.
Step 3: calculate interaction energies between titratable groups
The interaction energies between all titratable groups and other “fixed” charges in the molecule are required to calculate the titration curves in the next step. The finite difference Poisson-Boltzmann (FDPB) continuum electrostatics methodology (11,31), implemented in MEAD (32), is used to calculate the energetics of proton transfer. See Gordon et al. (25) for additional details.
A key input to the FDPB computations are the atomic partial charges in the protonated and deprotonated states of titratable groups. The partial charges from the AMBER force field parameters are used for the protonated and deprotonated states of ASP, CYS, GLU, HIS, LYS, and the deprotonated state of ARG and TYR. Since AMBER does not contain parameters for the rarely occurring protonated (neutral) state of ARG and TYR, these partial charges were obtained from Ryde (33) and MEAD, respectively.
H++ uses the single-conformer version of MEAD. However, conformational variability is partially accounted for by the ‘smeared charge’ representation of titratable groups, and the optimization of the positions of added protons. In the case of ASP and GLU, the charge of the additional proton in the protonated state is equally divided between the two O atoms to which it may be attached. In the case of ARG, HIS and LYS, the charge reduction in the deprotonated state is equally divided between the protons that may be removed. While not the most systematic or exhaustive way of incorporating conformational variability, we believe, based on previous experiences (18,31,34,35), that this particular model is a reasonable balance between speed and accuracy, and is therefore a good choice for a web-based calculation.
Step 4: calculate titration curves
The titration curve for a titratable group is the protonation probability as a function of solution pH. The protonation probability can be calculated as the Boltzmann average over the protonation states for the set of interaction energies calculated above (11). However, the exact computation of these statistical averages involves summations over all possible 2N microstates, where N is the number of titratable groups. To make the computation of Boltzmann averages tractable, two different approximations are used. For structures with less than 80 titratable groups, a clustering algorithm is employed to approximate the Boltzmann averages (36,37). For larger structures, the full Monte Carlo method as implemented in MCTI (12), is used. The pK of each group is computed as the mid-point of the corresponding titration curve, that is pK = pK1/2. Users are encouraged to check shapes of titration curves displayed by H++, as pK1/2 loses its meaning for titration curves that strongly deviate from the standard sigmoidal shape (38).
Step 5: generating outputs
H++ produces several outputs useful for molecular modeling and simulations. These include the estimated pK value for each titratable group, a PDB and the corresponding PQR file in the predicted protonation state, AMBER format topology and force field parameter files for explicit and implicit solvent simulations, energies of protonation microstates (for structures with <25 titratable groups), breakdown of contribution to pK shift of each group and the computed isoelectric point.
For efficiency, the titration curve is only calculated in the experimentally accessible pH range from 0 to 12. In some cases, the pH value at which the calculated protonation probability is 0.5 may be outside this range. For cases where the 0.5 protonation probability occurs outside the above pH range, the pK value is reported as ‘<0’ or ‘>12’.
The input PDB file is updated with titratable groups in their predicted protonation states based on estimated pK values and the user specified pH value—protonated if pH<pK, deprotonated otherwise. The PDB file is updated using the LEAP module in AmberTools which also adds atomic radii and partial charges producing a PQR format file. The LEAP module also produces AMBER force field parameter and coordinate files for running molecular dynamics simulations using AMBER or NAMD. These files can optionally include a cubic or octahedral solvent box with the user-specified number and type of (commonly used) ions, for explicit solvent simulations.
RESULTS
The accuracy of H++ was evaluated for a set of 23 high-quality structures (X-ray structures with resolution ≤2.50 A and no missing residues in the middle of the protein sequence), for which experimentally determined pK values are available (39) (Table 1). The set includes 201 titratable groups, of which 66 have large pK shifts (ΔpK ≥1.0) and 135 have small pK shifts (ΔpK <1.0). For comparison, predictions of a ‘Null’ model are also shown. Additional details for the results shown in Table 1, including PDB codes of the test proteins, can be found at http://biophysics.cs.vt.edu/faq.php.
Table 1.
RMS error in pK values computed by H++ relative to experiment, based on 23 protein structures with a total of 201 titratable groups
| ΔpK | RMS error |
Prot. accuracy (%) at pH 7 |
ΔpK direction accuracy (%) |
|||
|---|---|---|---|---|---|---|
| H++ | Null model | H++ | Null model | H++ | Null model | |
| ≥1.0 | 1.44 | 2.05 | 98 | 88 | 92 | 50 |
| <1.0 | 1.39 | 0.51 | 97 | 95 | 75 | 50 |
Results are broken down into two sets. One with large (>1.0) pK shifts (66 groups) and the other with small (≤1.0) pK shifts (135 groups). Table shows the RMS error, accuracy of predicted protonation at pH 7.0 and accuracy of predicted direction of pK shift, compared to experimentally determined values. The Null model assumes a pK shift of 0 from the model compound pK for the purpose of computing RMS error and predicted protonation state. For direction of the predicted pK shift the Null model assumes equal probabilities of ± shift.
Most of the test sets used for benchmarking pK prediction methods, including the one described above, lack groups with extreme pK shifts. To address this deficiency, we added to our test set a membrane-embedded protein bacteriorhodopsin (BR), a system with functionally relevant extreme pK shifts (40–42) (Table 2). The trends of the predicted pK shifts are in reasonable agreement with experiment. However, for the deeply buried groups, the use of a protein dielectric constant of 4, lower than the default value ϵin = 6, results in better agreement with experiment.
Table 2.
Experimental and computed pK values of the membrane-embedded Bacteriorhodopsin
| Site | Experiment |
H++ |
|
|---|---|---|---|
| ϵin = 6 (default) | ϵin = 4 | ||
| ARG 82 | >13.8 | >12 | >12 |
| ASP 85 | 2.6 | 2.3 | 1.8 |
| ASP 86 | >12 | 7.6 | >12 |
| ASP 115 | >9.5 | 8.0 | 10.4 |
| ASP 212 | <2.5 | <0 | <0 |
| Schiff Base (216) | >12 | >12 | >12 |
We used two values of protein dielectric (ϵin) constant. The structure corresponds to the BR resting state of the photocycle, and is constructed from PDB ID 1QHJ, as described in Onufriev et al. (34).
There are many factors that limit the accuracy of pK predicted by H++. Arguably, the more important factors are: shortcomings of the semi-microscopic, 2-dielectric PB implicit solvent model; incomplete accounting of conformational flexibility; and the absence of coupling between protonation and conformational states. Additional factors that affect the accuracy of pK prediction in general, can be found in (20) and (43). The PB implicit solvent model in H++ uses a fixed dielectric value for the solute (protein), even though the appropriate dielectric value can vary considerably depending on the location of the group in question within the protein (16,20,27,44,45). Significant improvement in pK prediction have also been reported when conformational flexibility is taken into account (46,47). Moreover, constant pH molecular dynamics studies have shown a strong correlation between conformation and protonation states (48,49). Inclusion of variable dielectric, conformational flexibility and the coupling between protonation states using currently available methods involve significant additional computational costs, which is counter to our goal of a fast web-based application.
CONCLUSION
The free open-source H++ web server described here automates the process of pK estimation and many of the steps required to prepare biomolecular structures for atomistic simulations. Input PDB structures are protonated according to the computed pKs, and force-field parameter/topology and coordinate files compatible with AMBER/NAMD formats are generated. The server is intended for both novice and expert users alike. A distinctive feature of H++ is that it includes extensive input consistency checks, safeguards, and pre- and post-processing warning messages that can help users make informed decisions when using the outputs from the server. The pK calculations in H++ are based on the established PB continuum electrostatics methodology that has been successfully used for this purpose for over two decades. The accuracy and usefulness of H++ has been improved in the latest version which includes several new capabilities: such as fixing erroneous (flipped) residue conformations; including a ligand in the input structure; processing nucleic acid structures; and generating a solvent box with specified number of common ions for explicit solvent MD. In addition, upgrades to the H++ server resulted in a 3× speedup relative to H++ 1.0 (25). We continue to enhance H++, with future plans to automate ligand processing and to add the ability to generate force field parameter/topology files for other popular modeling packages. Since its original release in August 2005, over 38 000 structures have been uploaded to H++ for processing by 2287 registered users and thousands of anonymous users, proving the usefulness of this tool to the molecular modeling and simulation community. The source code for H++ is also available, upon request, for offline batch processing of a large number of structures.
FUNDING
Funding open access charge: National Institutes of Health [GM076121 to A.V.O.].
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank John Gordon for helping to maintain the H++ server during the first 3 years of its operation.
REFERENCES
- 1.Dodson GG, Lane DP, Verma CS. Molecular simulations of protein dynamics: new windows on mechanisms in biology. EMBO Rep. 2008;9:144–150. doi: 10.1038/sj.embor.7401160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Freddolino PL, Arkhipov AS, Larson SB, McPherson A, Schulten K. Molecular dynamics simulations of the Complete Satellite Tobacco Mosaic Virus. Structure. 2006;14:437–449. doi: 10.1016/j.str.2005.11.014. [DOI] [PubMed] [Google Scholar]
- 3.Karplus M, McCammon JA. Molecular dynamics simulations of biomolecules. Nat. Struct. Biol. 2002;9:646–652. doi: 10.1038/nsb0902-646. [DOI] [PubMed] [Google Scholar]
- 4.Karplus M, Kuriyan J. Molecular dynamics and protein function. Proc. Natl. Acad. Sci. U.S.A. 2005;102:6679–6685. doi: 10.1073/pnas.0408930102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang W, Donini O, Reyes CM, Kollman PA. Biomolecular simulations: Recent developments in force fields, simulations of enzyme catalysis, protein-ligand, protein-protein, and protein-nucleic acid noncovalent interactions. Annu. Rev. Biophys. Biomol. Struct. 2001;30:211–243. doi: 10.1146/annurev.biophys.30.1.211. [DOI] [PubMed] [Google Scholar]
- 6.Schlick T, Guevara RC, Halvorsen LA, Jung S, Xiao X. Biomolecular modeling and simulation: A field coming of age. Quart. Rev. Biophys. 2011;44:191–228. doi: 10.1017/S0033583510000284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garcia-Viloca M, Poulsen TD, Truhlar DG, Gao J. Sensitivity of molecular dynamics simulations to the choice of the X-ray structure used to model an enzymatic reaction. Protein Science. 2004;13:2341–2354. doi: 10.1110/ps.03504104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Research. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tanford C, Kirkwood J. Theory of Protein Titration Curves. J. Am. Chem. Soc. 1957;79:5333–5339. [Google Scholar]
- 10.Tanford C, Roxby R. Interpretation of Protein Titration Curves. Biochemistry. 1972;11:2192–2198. doi: 10.1021/bi00761a029. [DOI] [PubMed] [Google Scholar]
- 11.Bashford D, Karplus M. pKa's of ionizable groups in proteins: Atomic detail from a continuum electrostatic model. Biochemistry. 1990;29:10219–10225. doi: 10.1021/bi00496a010. [DOI] [PubMed] [Google Scholar]
- 12.Beroza P, Fredkin DR, Okamura MY, Feher G. Protonation of interacting residues in a protein by Monte Carlo method. Proc. Natl. Acad. Sci. U.S.A. 1991;88:5804–5808. doi: 10.1073/pnas.88.13.5804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takahashi T, Nakamura H, Walda A. Electrostatic forces in two lysozymes: Calculations and measurements of histidine pKavalues. Biopolymers. 1992;32:897–909. doi: 10.1002/bip.360320802. [DOI] [PubMed] [Google Scholar]
- 14.Yang A-S, Gunner MR, Sampogna R, Sharp K, Honig B. On the Calculation of pKa's in Proteins. Proteins. 1993;15:252–265. doi: 10.1002/prot.340150304. [DOI] [PubMed] [Google Scholar]
- 15.DelBuono G, Figueirido F, Levy R. Intrinsic pKas of ionizable residues in proteins: An explicit solvent calculation for lysozyme. Proteins. 1994;20:85–97. doi: 10.1002/prot.340200109. [DOI] [PubMed] [Google Scholar]
- 16.Demchuk E, Wade RC. Improving the continuum dielectric approach to calculating pKa's of ionizable groups in proteins. J. Phys. Chem. 1996;100:17373–17387. [Google Scholar]
- 17.Sham YY, Chu ZT, Warshel A. Consistent Calculations of pKa's of Ionizable Residues in Proteins: Semi-microscopic and Microscopic Approaches. J. Phys. Chem. 1997;101:4458–4472. [Google Scholar]
- 18.Ullmann GM, Knapp E-W. Electrostatic models for computing protonation and redox equilibria in proteins. Eur.Biophys.J. 1999;28:533–551. doi: 10.1007/s002490050236. [DOI] [PubMed] [Google Scholar]
- 19.Spassov VZ, Bashford D. Multiple-Site Ligand Binding to Flexible Macromolecules. J. Comp. Chem. 1999;20:1091–1111. [Google Scholar]
- 20.Antosiewicz J, McCammon JA, Gilson MK. Prediction of pH-dependent Properties of Proteins. J. Mol. Biol. 1994;238:415–436. doi: 10.1006/jmbi.1994.1301. [DOI] [PubMed] [Google Scholar]
- 21.Nielsen JE, Vriend G. Optimizing the hydrogen-bond network in Poisson-Boltzmann equation-based pKa calculations. Proteins. 2001;43:403–412. doi: 10.1002/prot.1053. [DOI] [PubMed] [Google Scholar]
- 22.Georgescu R, Alexov E, Gunner M. Combining Conformational Flexibility and Continuum Electrostatics for Calculating pKas in Proteins. Biophysical Journal. 2002;83:1731–1748. doi: 10.1016/S0006-3495(02)73940-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mongan J, Case DA, McCammon JA. Constant pH Molecular Dynamics in Generalized Born Implicit Solvent. J. Comp. Chem. 2004;25:2038–48. doi: 10.1002/jcc.20139. [DOI] [PubMed] [Google Scholar]
- 24.Baker NA, Bashford D, Case DA. Implicit solvent electrostatics in biomolecular simulation. In: Leimkuhler B, Chipot C, Elber R, Laaksonen A, Mark A, Schlick T, Schütte C, Skeel R, editors. New Algorithms for Macromolecular Simulation. Vol. 49. Springer Berlin/Heidelberg; 2006. pp. 263–295. Chapter 15. [Google Scholar]
- 25.Gordon JC, Myers JB, Folta T, Shoja V, Heath LS, Onufriev AV. H++: a server for estimating pKa's and adding missing hydrogens to macromolecules. Nucleic Acids Res. 2005;33:68–71. doi: 10.1093/nar/gki464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Word MJ, Lovell SC, Richardson JS, Richardson DC. Asparagine and glutamine: using hydrogen atom contacts in the choice of side-chain amide orientation. J. Mol. Biol. 1999;285:1735–1747. doi: 10.1006/jmbi.1998.2401. [DOI] [PubMed] [Google Scholar]
- 27.Harms MJ, Schlessman JL, Chimenti MS, Sue GR, Damjanović A, García-Moreno B. A buried lysine that titrates with a normal pKa: role of conformational flexibility at the protein-water interface as a determinant of pKa values. Protein sci. 2008;17:833–845. doi: 10.1110/ps.073397708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O'Boyle N, Banck M, James C, Morley C, Vandermeersch T, Hutchison G. Open Babel: An open chemical toolbox. J. Cheminformat. 2011;3:33. doi: 10.1186/1758-2946-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Case DA, Cheatham TE, Darden T, Gohlke H, Luo R, Merz KM, Onufriev A, Simmerling C, Wang B, Woods RJ. The Amber biomolecular simulation programs. J. Comp. Chem. 2005;26:1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bondi A. van der Waals Volumes and Radii. J. Phys. Chem. 1964;68:441–451. [Google Scholar]
- 31.Bashford D, Gerwert K. Electrostatic Calculations of the pKa Values of Ionizable Groups in Bacteriorhodopsin. J. Mol. Biol. 1992;224:473–486. doi: 10.1016/0022-2836(92)91009-e. [DOI] [PubMed] [Google Scholar]
- 32.Bashford D. Vol. 1343. Springer Berlin / Heidelberg; 1997. An object-oriented programming suite electrostatic effects in biological molecules. An experience report on the MEAD project. Scientific Computing in Object-Oriented Parallel Environments. of Lecture Notes in Computer Science, chapter 30 pp. 233–240. [Google Scholar]
- 33.Sigfridsson E, Ryde U. Comparison of methods for deriving atomic charges from the electrostatic potential and moments. J. Comput. Chem. 1998;19:377–395. [Google Scholar]
- 34.Onufriev A, Smondyrev A, Bashford D. Proton Affinity Changes During Unidirectional Proton Transport in the Bacteriorhodopsin Photocycle. J. Mol. Biol. 2003;332:1183–1193. doi: 10.1016/s0022-2836(03)00903-3. [DOI] [PubMed] [Google Scholar]
- 35.Spassov VZ, Yan L. A fast and accurate computational approach to protein ionization. Protein Sci. 2008;17:1955–1970. doi: 10.1110/ps.036335.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Anandakrishnan R, Onufriev A. Analysis of Basic Clustering Algorithms for Numerical Estimation of Statistical Averages in Biomolecules. J. Comput. Biol. 2008;15:165–184. doi: 10.1089/cmb.2007.0144. [DOI] [PubMed] [Google Scholar]
- 37.Myers J, Grothaus G, Narayanan S, Onufriev A. A simple clustering algorithm can be accurate enough for use in calculations of pKs in macromolecules. Proteins. 2006;63:928–938. doi: 10.1002/prot.20922. [DOI] [PubMed] [Google Scholar]
- 38.Onufriev A, Case DA, Ullmann GM. A novel view of pH titration in biomolecules. Biochemistry. 2001;40:3413–3419. doi: 10.1021/bi002740q. [DOI] [PubMed] [Google Scholar]
- 39.Grimsley GR, Scholtz JM, Pace CN. A summary of the measured pK values of the ionizable groups in folded proteins. Prot. Sci. 2009;18:247–251. doi: 10.1002/pro.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Spassov VZ, Luecke H, Gerwert K, Bashford D. pKa calculations suggest storage of an excess proton in a hydrogen-bonded water network in bacteriorhodopsin. J. Mol. Biol. 2001;312:203–219. doi: 10.1006/jmbi.2001.4902. [DOI] [PubMed] [Google Scholar]
- 41.Kono M, Misra S, Ebrey TG. pH dependence of light-induced proton release by bacteriorhodopsin. FEBS Lett. 1993;331:31–34. doi: 10.1016/0014-5793(93)80291-2. [DOI] [PubMed] [Google Scholar]
- 42.Balashov SP, Imasheva ES, Govindjee R, Ebrey TG. Titration of aspartate-85 in bacteriorhodopsin: what it says about chromophore isomerization and proton release. Biophys. J. 1996;70:473–481. doi: 10.1016/S0006-3495(96)79591-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Castañeda CA, Fitch CA, Majumdar A, Khangulov V, Schlessman JL, García-Moreno BE. Molecular determinants of the pKa values of Asp and Glu residues in staphylococcal nuclease. Proteins. 2009;77:570–588. doi: 10.1002/prot.22470. [DOI] [PubMed] [Google Scholar]
- 44.Schutz CN, Warshel A. What are the dielectric constants of proteins and how to validate electrostatic models? Proteins. 2001;44:400–417. doi: 10.1002/prot.1106. [DOI] [PubMed] [Google Scholar]
- 45.Isom DG, Castañeda CA, Cannon BR, Velu PD, García-Moreno EB. Charges in the hydrophobic interior of proteins. Proc. Natl Acad. Sci. USA. 2010;107:16096–16100. doi: 10.1073/pnas.1004213107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Song Y, Mao J, Gunner MR. MCCE2: Improving protein pKa calculations with extensive side chain rotamer sampling. J. Comput. Chem. 2009;30:2231–2247. doi: 10.1002/jcc.21222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Powers N, Jensen JH. Chemically accurate protein structures: Validation of protein NMR structures by comparison of measured and predicted pKa values. J. Biomol. NMR. 2006;35:39–51. doi: 10.1007/s10858-006-9003-3. [DOI] [PubMed] [Google Scholar]
- 48.Lee MS, Salsbury FR, Brooks CL. Constant-pH molecular dynamics using continuous titration coordinates. Proteins. 2004;56:738–752. doi: 10.1002/prot.20128. [DOI] [PubMed] [Google Scholar]
- 49.Mongan J, Case DA, McCammon JA. Constant pH molecular dynamics in generalized Born implicit solvent. J. Comput. Chem. 2004;25:2038–2048. doi: 10.1002/jcc.20139. [DOI] [PubMed] [Google Scholar]