Abstract
Since its first release in 2007, GeneCodis has become a valuable tool to functionally interpret results from experimental techniques in genomics. This web-based application integrates different sources of information to finding groups of genes with similar biological meaning. This process, known as enrichment analysis, is essential in the interpretation of high-throughput experiments. The frequent feedbacks and the natural evolution of genomics and bioinformatics have allowed the growth of the tool and the development of this third release. In this version, a special effort has been made to remove noisy and redundant output from the enrichment results with the inclusion of a recently reported algorithm that summarizes significantly enriched terms and generates functionally coherent modules of genes and terms. A new comparative analysis has been added to allow the differential analysis of gene sets. To expand the scope of the application, new sources of biological information have been included, such as genetic diseases, drugs–genes interactions and Pubmed information among others. Finally, the graphic section has been renewed with the inclusion of new interactive graphics and filtering options. The application is freely available at http://genecodis.cnb.csic.es.
INTRODUCTION
Next-generation sequencing and other high-throughput experimental techniques such as DNA microarrays are revolutionary technologies for advanced biomedical research. Instead of studying individual genes or proteins, these techniques tackle the study of different experimental conditions from a global perspective. A large list of genes or proteins is usually resulted after processing the massive amount of information generated by these techniques. The interpretation of these lists in a biological context is the next logical step.
Several data mining techniques have been developed for a better understanding of the experiments. One of the most popular methods consists in finding biological annotations statistically enriched in lists of genes or proteins compared with a reference set, which is often the entire genome or the complete list of genes in a study. Tools that translate large lists of genes or proteins into related terms from Gene Ontology (GO) (1) such as Onto-Express (2), DAVID (3), FATIGO+ (4) or ProfCom (5) are now routinely used. A good compendium of available tools can be found at http://www.geneontology.org/GO.tools.shtml (accessed April 30, 2012).
According to a recent review (6), current functional enrichment algorithms can be classified into three groups: singular enrichment analysis (SEA), gene set enrichment analysis (GSEA) and modular enrichment analysis (MEA). SEA is probably the most typical way to make an enrichment analysis (7,8). This strategy looks for individual annotations related to genes or proteins in different biological databases and evaluates the statistical significance of each individual term. This is commonly made by counting the number of genes from the input and reference lists containing the term and computing a statistical test, usually χ2 or Fisher's test and using Binomial or Hypergeometric distributions. The computed P-value represents the significance of the association between the term and the input list of molecules.
GSEA (9) works in the same way as SEA but taking into account the whole list of genes instead of analyzing the most relevant ones. Its main goal is to avoid the arbitrary cutoff in expression level. The main idea is to know if the set of genes related with a specific term tends to concentrate at the top (the most overexpressed) or at the bottom (the most inhibited) of the list. There are few available tools for GSEA like FatiScan (10) or GO-Mapper (11).
Although these strategies have been demonstrated to be useful, the lack of term–term relationships in their analyses is an important drawback. The combinations of terms extend our understanding of biological events associated with a given experimental system. It provides a more specific meaning while increasing the quality of results by detecting additional significant terms. As shown in (12), there are biological terms not detected as statistically relevant when tested individually, which turn out to be significant when appearing together with others. MEA methods avoid this problem and provide a more complete functional analysis. Two examples of MEA tools are DAVID, that does clustering of singular enrichment results and ProfCom, that takes into account different kinds of logic relationships between the terms (AND, OR, NOT).
In 2007, we introduced GeneCodis (12,13), a tool for singular and modular enrichment analysis that integrates information of diverse nature (e.g. functional, regulatory or structural) by looking for frequent patterns in the space of annotations and computing their statistical relevance. This integrative capacity sheds light on different aspects of the same information and provides a more accurate interpretation of the data.
In this work, we present a new release of GeneCodis with several novel features. New types of annotations now expand its analysis capacity. For example, Pubmed Ids, pathways from Panther (14) or drug target genes from PharmGKB (15). Exome analysis is also possible by the inclusion of exon ids from Ensembl (16). Another significant novelty consists in comparing different lists of genes or proteins (17,18), focused in finding shared and divergent information related with different experimental conditions. GeneCodis3 also integrates a new post-analysis algorithm to remove the noise and the intrinsic redundancy of the biological annotations. Finally, a new graphical section has been implemented, with new interactive visualizations and the option to filter and highlight terms.
NEW FEATURES
The standard workflow of the tool has not been drastically modified to keep the same logic and consistency with the previous version. The analysis is made by pasting or loading a list of genes and selecting the organism and the categories of annotations. The job is sent to our servers, and once the results are ready, all visualizations are displayed. MEA and SEA are always simultaneously executed for every submission. Keeping the web browser opened while executing the job is not necessary. Results can be retrieved by keeping the active URL or by introducing an email address to receive a notification when the job is ready. The new features, explained more deeply below, are intended to facilitate the interpretation of the results and to make the system more self-contained. Table 1 summarizes the differences between previous and current versions.
Table 1.
Comparative table between previous and current versions of GeneCodis
| Features | GeneCodis2 | GeneCodis3 |
|---|---|---|
| Singular enrichment analysis | ✓ | ✓ |
| Modular enrichment analysis | ✓ | ✓ |
| Comparative analysis | ✓ | |
| Redundancy removal (GTLinker) | ✓ | |
| Functional annotations | ✓ | ✓ |
| Regulatory annotations | ✓ | ✓ |
| Pharmacological annotations | ✓ | |
| Disease annotations | ✓ | |
| Panther pathways annotations | ✓ | |
| Pubmed annotations | ✓ | |
| Graphics section | ✓ | ✓ |
| Interactive visualization | ✓ | |
| Tag cloud of terms | ✓ | |
| Computing cluster support | ✓ | ✓ |
| Multithreading support | ✓ |
Non-redundant reciprocal linkage of genes and terms
Enrichment analysis techniques are very useful to extract biological knowledge from a set of genes or proteins, but at the same time, they frequently generate a long list of significant terms, even for a very small set of molecules. The problem is worse when different annotations are used because the size of the problem increases significantly.
Most of these large sets of results suffer from the redundancy of the biological terms that are repeated in many different annotation resources. For example, GO, KEGG (19) and others have different terms to express the same event (e.g. Glycolysis GO:0006096 in GO and Glycolysis/Glucogenesis 00010 pathway in KEGG). The GO hierarchy also affects the results since some similar terms are used to express the same reality (e.g. GO:0065007 biological regulation and GO:0050789 regulation of biological process). Additionally, the bias in the annotation space also affects the enrichment since there are frequent general terms that do not provide very useful information, such as the GO:0007049 cell cycle term.
A recently proposed computational method, called GeneTerm Linker or GTLinker for short (20), summarizes the enrichment results by finding significant and coherent collections of genes and terms. This approach executes several filtering and clustering steps to eliminate redundant and non-informative terms and produces genes and annotations grouped in modules (metagroups). These metagroups, which are supposed to be functionally coherent, absorb the redundancy of the original significant terms and are ranked by their significance and coherence.
We have implemented this method in GeneCodis as an optional post-analysis of the results. The output contains a table with the cluster of genes and annotations as well as their statistical significance. Details can also be displayed to show the original sets of the enrichment analysis, and the similarity coefficient of the resulting group. Currently, only Homo sapiens and Saccharomyces cerevisiae are supported, but the rest of organisms will be incrementally added.
Comparative analysis
It is very common in high-throughput experiments to work with different conditions that finally derive in different groups of genes. In these cases, the challenge consists in interpreting the functional differences of the groups. Executing an enrichment analysis for each set of genes and manually comparing them is the traditional way to do it. This time-consuming task could be difficult when working with large lists.
We have now included a comparative analysis tool that allows the submission of two different lists at the same time and carries out a simultaneous modular and singular enrichment analyses for each one. The most valuable information when comparing two lists is the common and divergent information. Three additional analyses are performed for the intersection of the two sets as well as the original ones without the intersecting area. These operations are done over the space of genes instead of the annotations to guarantee the statistical significance of the results.
The results are shown in a similar way to the results of a standard analysis. For each category, an interactive Venn diagram with the number of genes of each group is displayed. When clicking on each group of the diagram, a table with its corresponding results is opened. Results for the original sets are also included.
New sources of annotations
Due to the highly distributed nature of biological information, one of the most important tasks of GeneCodis consists in the integration of all the data in a unique local resource to allow an assortment of analyses. We have used Biomart (21) to automate these data accession and management tasks.
GeneCodis already contains an extensive amount of information from different resources. For example, GO, KEGG pathways, structural information via the InterPro protein domains (22) and regulatory information from microRNAs targets from mirBase (23). Transcription factors (TFs) were extracted from curated sources. For human, rat and mouse, TFs were extracted from the MsigDB database (9), mainly based on TransFac (24). In the case of yeast, TFs were obtained from the experimental data contained in YEASTRACT database (25).
This release supports gene-related diseases from the online Mendelian inheritance in man (OMIM) database (26), which contains complete information about the known mendelian disorders. Information from PharmGKB is included to measure the effect of genetic activity in response to drugs. This annotation increases its value when considered together with OMIM. Panther is another ontology-based database of pathways (mainly signaling pathways) that, jointly with KEGG, provides an excellent resource to know the map of interactions of a given gene list. The inclusion of biomedical literature data from Pubmed identifiers (http://www.ncbi.nlm.nih.gov/pubmed/, accessed April 30, 2012) facilitates the finding of articles related with a group of genes. Finally, in consonance with the recent increase of exome resequencing studies, GeneCodis accepts exons identifiers from Ensembl to find the significant terms that are overrepresented in the genes composed by any of the input exons.
User interface
When analyzing big sets of genes, results are usually too large for visual analysis. Graphics such as pie or column charts are very useful to quickly detect the most significant combinations of terms. New interactive graphics have been integrated in this version, as well as filters to select the groups of annotations, either by name or by number of genes.
GeneCodis also includes a new tag cloud that displays the 30 most significant terms of the results. The tag’s sizes vary according to the number of supporting genes and are also linked to their corresponding databases. This graphical representation allows a rapid interpretation of the most relevant content of the analysis.
Like in the previous release, GeneCodis supports programmatically access through the SOAP Web Services technology. With a few lines of code, GeneCodis can be integrated in any kind of pipeline, regardless of the programming language or platform.
GENECODIS USE CASE AND OUTPUT DESCRIPTION
We demonstrate the utility of GeneCodis by analyzing the differentially expressed genes of human lung WI-38 fibroblasts upon exposure to the ultimate carcinogen benzo[a]pyrene diol epoxide (BPDE) (17). In this study, normal human WI-38 lung fibroblasts were exposed to three different concentrations of BPDE, and whole-genome oligonucleotide microarrays were used to measure changes in gene expression. WI-38 cells were exposed to 0.1, 0.5 or 1 µM BPDE for 24 h which resulted in 384, 972 and 837 differentially expressed genes, respectively. Results of this study are summarized in Figure 1 of (17).
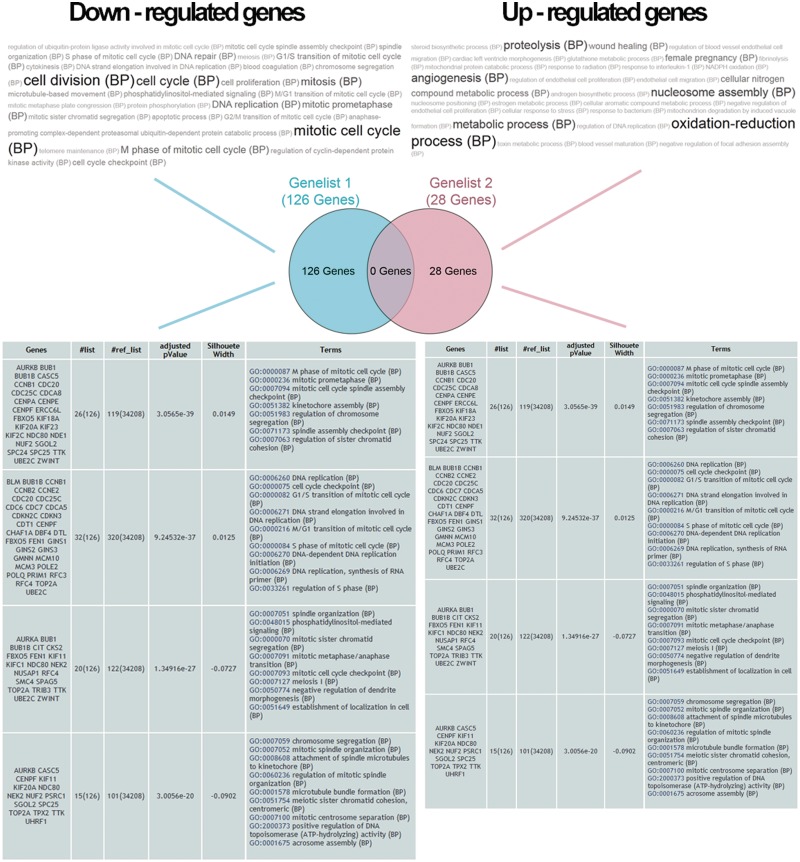
Figure 1.
Collage of output generated by GeneCodis. The Venn diagram contains the number of actual genes in each list after annotation. Two tag cloud visualizations containing significant enriched terms in each analysis are shown on the top. The two tables on the bottom show the four most relevant metagroups of genes and terms generated after running the integrated GTLinker on each list to avoid redundancy.
Using the same list of genes that were reported as expressed in response to all doses of BPDE, we repeated the analysis with GeneCodis using the Comparative option, which is very convenient for experimental designs involving more than one condition. Results are shown in Figure 1. As input we submitted two lists of genes, the organism (human in this case) and the annotations. To reproduce the results in the article, we only used GO Biological Process.
The central part of Figure 1 shows a Venn diagram containing the number of genes in each list after annotation. Genes with no annotations are removed and not considered in the statistics. The top of the figure shows the cloud of tags that contains significant enriched terms in each analysis. A simple visual inspection already provides information about the functional implication of the gene sets. As explained in (17), downregulated genes are involved in mitosis, cell cycle, spindle organization and biogenesis as well as DNA replication and DNA repair. Similarly, biological processes enriched in upregulated genes affected by all doses of BPDE are highlighted in the right cloud of tags and show the involvement of all these genes in the nucleosome assembly, cell proliferation and extracellular processes.
The bottom of Figure 1 shows the metagroups generated after running the integrated GTLinker to avoid redundancy in the original enrichment results. In the case of repression, GeneCodis generated 203 statistical significant terms and GTLinker clustered them into a reduced set of 41 metagroups. The left table in Figure 1 shows the top four metagroups according to their significance (P-value). In a similar fashion, the table in the right shows the top four metagroups out of a total of eight metagroups that were generated from the 57 enrichment results of the induced genes. By exploring the GO terms in each metagroup, we can identify the main biological processes that are related to each condition. The coherence and succinctness of the information provided by the most significant metagroups, together with the comparative functionality and visualizations, have made the reproducibility of this study quite simple and automatic.
TECHNICAL DETAILS
GeneCodis core consists in a C++ code which includes a method to generate combination of frequent biological terms that are present in the gene sets (27). A Ruby wrapper processes all data and creates a script to run the core program, balancing the computational load by deciding which processor will execute the job. The Ruby wrapper has been improved from the previous versions by taking advantage of new releases of this language, making use of threads to re-implement potentially parallel parts of the program.
Client–server architecture has been used to decouple the front-end from the heavy computational code that runs on a different server. We use a cluster of six new-generation servers, each one containing two Quad-Core Intel Xeon 64 bits processors, that is able to handle a large number of simultaneous jobs. The web portal has been ported to Ruby on Rails, a well-tested and stable technology. As in the previous version, the whole functionality is also supported by web services to allow programmatic access to the tool.
This new version has been running for a few months, experiencing more than 1000 submissions. Extensive tests have been carried out in different web browsers using synthetic and real data sets for which the outcome of the software is known.
DISCUSSION
Since the launch of its second release, GeneCodis received more than 43 500 effective visits, each of them executing several jobs, which gives an indication of the popularity of the tool. One of the disadvantages of enrichment analysis and more in particular with the modular enrichment is the generation of hundreds of significant results loaded with an inherent redundancy that make interpretation very difficult. This is caused by the natural redundancy of the information across biological databases. In this new release, we have tackled this issue with the inclusion of a recently reported algorithm, which groups and filters the data extracted from the enrichment analysis. This is complemented with the addition of several new sources of information.
The comparative functionality also represents a qualitative improvement. It allows an easy comparison of gene sets in the same step by executing simultaneous analyses of their possible combinations. This is in concert with the common experimental designs in high-throughput experiments. The new graphical section also improves the way to visualize the results, allowing a direct interaction with the charts and tables. A special attention should be given to the cloud of tags, which produces a quick snapshot of the most prominent and significant enriched terms. Even if this visualization was not intended to summarize the information, it acts effectively as such.
The new functionality, extensively described in this article, together with the new visualization and computational infrastructure, makes GeneCodis a novel functional genomics tool compared not only with its previous versions but also with some of the other existing tools in the field.
FUNDING
Spanish Minister of Science and Innovation [BIO2010-17527]; Government of Madrid (CAM) [P2010/BMD-2305]; Juan de la Cierva research program (to R.N.-C.). Funding for open access charge: Spanish Minister of Science and Innovation [BIO2010-17527].
Conflict of interest statement. None declared.
REFERENCES
- 1.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. The Gene Ontology Consortium Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nature genetics. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Khatri P, Draghici S, Ostermeier GC, Krawetz SA. Profiling gene expression using onto-express. Genomics. 2002;79:266–270. doi: 10.1006/geno.2002.6698. [DOI] [PubMed] [Google Scholar]
- 3.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 4.Al-Shahrour F, Minguez P, Tárraga J, Medina I, Alloza E, Montaner D, Dopazo J. FatiGO +: a functional profiling tool for genomic data. Integration of functional annotation, regulatory motifs and interaction data with microarray experiments. Nucleic Acids Res. 2007;35:W91–W96. doi: 10.1093/nar/gkm260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Antonov AV, Schmidt T, Wang Y, Mewes HW. ProfCom: a web tool for profiling the complex functionality of gene groups identified from high-throughput data. Nucleic Acids Res. 2008;36:W347–W351. doi: 10.1093/nar/gkn239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Khatri P, Drăghici S. Ontological analysis of gene expression data: current tools, limitations, and open problems. Bioinformatics. 2005;21:3587–3595. doi: 10.1093/bioinformatics/bti565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dopazo J. Functional interpretation of microarray experiments. Omics. 2006;10:398–410. doi: 10.1089/omi.2006.10.398. [DOI] [PubMed] [Google Scholar]
- 9.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Al-Shahrour F, Arbiza L, Dopazo H, Huerta-Cepas J, Mínguez P, Montaner D, Dopazo J. From genes to functional classes in the study of biological systems. BMC Bioinformatics. 2007;8:114. doi: 10.1186/1471-2105-8-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smid M, Dorssers LCJ. GO-Mapper: functional analysis of gene expression data using the expression level as a score to evaluate Gene Ontology terms. Bioinformatics. 2004;20:2618–2625. doi: 10.1093/bioinformatics/bth293. [DOI] [PubMed] [Google Scholar]
- 12.Carmona-Saez P, Chagoyen M, Tirado F, Carazo JM, Pascual-Montano A. GENECODIS: a web-based tool for finding significant concurrent annotations in gene lists. Genome Biol. 2007;8:R3. doi: 10.1186/gb-2007-8-1-r3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nogales-Cadenas R, Carmona-Saez P, Vazquez M, Vicente C, Yang X, Tirado F, Carazo JM, Pascual-Montano A. GeneCodis: interpreting gene lists through enrichment analysis and integration of diverse biological information. Nucleic Acids Res. 2009;37:W317–W322. doi: 10.1093/nar/gkp416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mi H, Dong Q, Muruganujan A, Gaudet P, Lewis S, Thomas PD. PANTHER version 7: improved phylogenetic trees, orthologs and collaboration with the Gene Ontology Consortium. Nucleic Acids Res. 2010;38:D204–D210. doi: 10.1093/nar/gkp1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McDonagh EM, Whirl-Carrillo M, Garten Y, Altman RB, Klein TE. From pharmacogenomic knowledge acquisition to clinical applications: the PharmGKB as a clinical pharmacogenomic biomarker resource. Biomark. Med. 2011;5:795–806. doi: 10.2217/bmm.11.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Flicek P, Amode MR, Barrell D, Beal K, Brent S, Chen Y, Clapham P, Coates G, Fairley S, Fitzgerald S, et al. Ensembl 2011. Nucleic Acids Res. 2011;39:D800–D806. doi: 10.1093/nar/gkq1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dreij K, Rhrissorrakrai K, Gunsalus KC, Geacintov NE, Scicchitano DA. Benzo[a]pyrene diol epoxide stimulates an inflammatory response in normal human lung fibroblasts through a p53 and JNK mediated pathway. Carcinogenesis. 2010;31:1149–1157. doi: 10.1093/carcin/bgq073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rezaul K, Thumar JK, Lundgren DH, Eng JK, Claffey KP, Wilson L, Han DK. Differential protein expression profiles in estrogen receptor-positive and -negative breast cancer tissues using label-free quantitative proteomics. Genes Cancer. 2010;1:251–271. doi: 10.1177/1947601910365896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2011;40:D109–D114. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fontanillo C, Nogales-Cadenas R, Pascual-Montano A, De Las Rivas J. Functional analysis beyond enrichment: non-redundant reciprocal linkage of genes and biological terms. PLoS One. 2011;6:e24289. doi: 10.1371/journal.pone.0024289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kasprzyk A. BioMart: driving a paradigm change in biological data management. Database. 2011 doi: 10.1093/database/bar049. November 11 (doi:10.1093/database/bar049; epub ahead of print). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hunter S, Jones P, Mitchell A, Apweiler R, Attwood TK, Bateman A, Bernard T, Binns D, Bork P, Burge S, et al. InterPro in 2011: new developments in the family and domain prediction database. Nucleic Acids Res. 2011;40:D306–D312. doi: 10.1093/nar/gkr948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39:D152–D157. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matys V. TRANSFAC(R): transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 2003;31:374–378. doi: 10.1093/nar/gkg108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abdulrehman D, Monteiro PT, Teixeira MC, Mira NP, Lourenço AB, dos Santos SC, Cabrito TR, Francisco AP, Madeira SC, Aires RS, et al. YEASTRACT: providing a programmatic access to curated transcriptional regulatory associations in Saccharomyces cerevisiae through a web services interface. Nucleic Acids Res. 2011;39:D136–D140. doi: 10.1093/nar/gkq964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hamosh A. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 2002;30:52–55. doi: 10.1093/nar/30.1.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Borgelt C, Yang X, Nogales-Cadenas R, Carmona-Saez P, Pascual-Montano A. Proceedings of the 14th International Conference on Extending Database Technology - EDBT/ICDT ’11. New York, USA: ACM Press; 2011. Finding closed frequent item sets by intersecting transactions; p. 367. [Google Scholar]