Abstract
EvolView is a web application for visualizing, annotating and managing phylogenetic trees. First, EvolView is a phylogenetic tree viewer and customization tool; it visualizes trees in various formats, customizes them through built-in functions that can link information from external datasets, and exports the customized results to publication-ready figures. Second, EvolView is a tree and dataset management tool: users can easily organize related trees into distinct projects, add new datasets to trees and edit and manage existing trees and datasets. To make EvolView easy to use, it is equipped with an intuitive user interface. With a free account, users can save data and manipulations on the EvolView server. EvolView is freely available at: http://www.evolgenius.info/evolview.html.
INTRODUCTION
In the context of current comparative genomics and metagenomics projects, graphic representations of phylogenetic trees go well beyond simple cladograms or phylograms. Trees are increasingly used to visualize comparative information in an evolutionary context, e.g. when comparing the fraction of genes involved in specific processes in each phylum. A wide variety of tree-viewing tools have been developed in the past years, such as TreeView (1), Dendroscope (2), iTol (3), jsPhyloSVG (4), PhyloWidget (5) and many others (6–16). However, none of these systems allows users to not only customize phylogenetic trees and associated annotation, but also manage both of them interactively (see Supplementary Table 1, which gives a brief overview over existing programs for tree visualization). We thus developed EvolView, an online web application that links any standard web browser to a personalized library of customized and annotated phylogenetic trees. EvolView is not only a visualization and customization tool, but also a management system for phylogenetic trees and associated datasets (annotations). Its carefully designed user interface makes using EvolView intuitive, simple and efficient.
FEATURES
A tree visualization and customization tool
EvolView is a comprehensive tool for tree visualization and annotation. It takes phylogenetic trees in several formats as input, including Newick/ Phylip, Nexus (17), Nhx and PhyloXML (18) formats. EvolView can display trees as phylograms and cladograms, each in either rectangular or circular layout.
Using the toolbar (Figure 1), users can easily change tree representation modes as well as visualization details such as leaf font size, branch line width and overall width and height of the trees. More complex customizations such as branch and leaf colors, leaf backgrounds or leaf-associated pie-charts and bar-plots are based on user-supplied datasets.
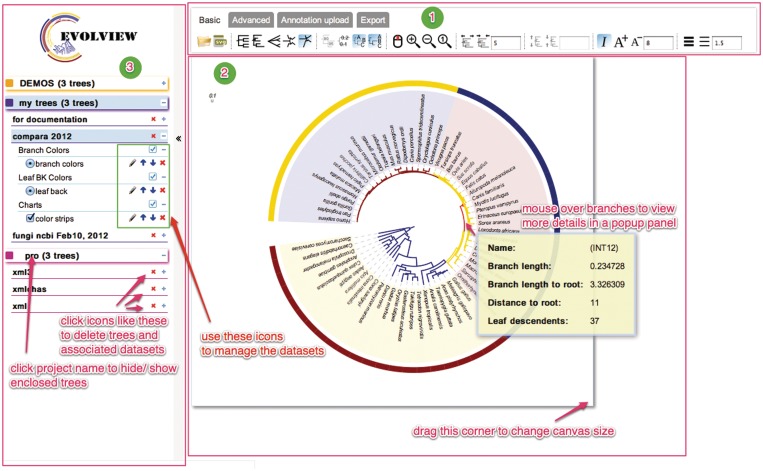
Figure 1.
The user interface of EvolView. Functions of some widgets are highlighted with red text. The interface consists of three parts. (i) A multi-tab toolbar contains sets of icons for tree and data manipulation, data upload and export into various graphical formats. (ii) An interactive canvas displays the annotated tree exactly as it will be reproduced in publication-ready image files. (iii) The browsing panel shows user data and provides widgets for data management.
The customized results can be exported into a wide range of graphical formats, including SVG, PDF, PNG, GIF and JPEG. Exporting phylogenetic trees to textual formats such as Newick, Nexus, Nhx and PhyloXML is also supported.
A management system for phylogenetic trees and associated datasets
EvolView also makes tree and dataset management easy. Trees are organized into user-defined ‘projects’ (Figure 1). Upon submission of a new tree, users are prompted to either create a new project, or to choose an existing project that groups related trees together.
With EvolView, users can easily add new annotation datasets to a tree, delete existing ones or edit their contents. Changing the display order of annotations on the tree and switching annotations on and off are also supported.
With a free account, all user data as well as tree customizations and data manipulations will be securely saved on the EvolView server. Thus, reopened trees appear exactly as they were left.
A comprehensive and intuitive user-interface
EvolView features a single-page interface (Figure 1) that facilitates a fluid user experience, where tree viewing, editing and managing are controlled on the same web page.
The EvolView interface consists mainly of three panels, as shown in Figure 1. (i) A multi-tab toolbar contains sets of clickable icons. Tooltips are provided when mousing over each icon. (ii) An interactive graphical editor that shows immediately all changes made by users. Users can zoom in and out of a plot, drag and drop the tree to a desired position, change the size of the canvas to fit the plot and export it to publication-ready graphical formats that reproduce exactly what is shown on the canvas. (iii) A hierarchical browsing panel lists all datasets of the current user and provides an interface for managing projects, trees and associated datasets. By default, a ‘DEMO’ project containing three trees is loaded; these trees were prepared to demonstrate the key features of EvolView.
A unique dataset system for graphical annotations
EvolView supports the usage of external datasets for customization. We expanded the dataset system so that it contains not only the data to be displayed on the trees, but also controlling commands we call ‘modifiers’ to change the default display options of the data. As shown in Table 1, the available modifiers cover almost all aspects of the customization, including, e.g. the plot area, opacity and minimal and maximal radiuses for pie charts. We also introduced a legend system based on modifiers, which can be used to choose the title, contents, colors and styles of each legend.
Table 1.
‘Modifiers’ supported by the dataset system
| Modifiers (case insensitive) | Value | Description |
|---|---|---|
| !groups or !LegendText | Comma separated text | Legend texts |
| !LegendStyle or !Style | rect/circle/star | Shapes to be plotted before the legends |
| !LegendColors or !Colors | Comma separated color codes or names | Colors to be applied to the shapes specified by LegendStyle |
| !Title or !Legend | Text | Title of the legend |
| !showLegends or !showLegend | 0/1 | 0: hide legends; 1: show legends |
| !opacity or !op | Any float number between 0 and 1 | Opacity of the graphical annotation |
| !plotWidth | Any integer > 0 | Graphical width of the dataset on the canvas |
| !minradiusa | Any float number > 0 | Minimal radius of the pies |
| !maxradiusa | Any float number > 0 | Maximal radius of the pies |
| !areaa | None | Use user inputs as the areas of the pies |
| !radiusa | None | Use user inputs as the radiuses of the pies |
| !gridb | None | Show grid lines behind the plots |
| !gridlabel or !axisb | None | Show numbers corresponding to the grid lines |
| !fanPlot or !fanb | None | Plot bars as fans in circular plot |
aFor pie charts only.
bfor bar charts only. For more details about the datasets and more examples, please see our online documentation on datasets at http://code.google.com/p/evolgenius/wiki/EvolViewIntroductionToDatasets.
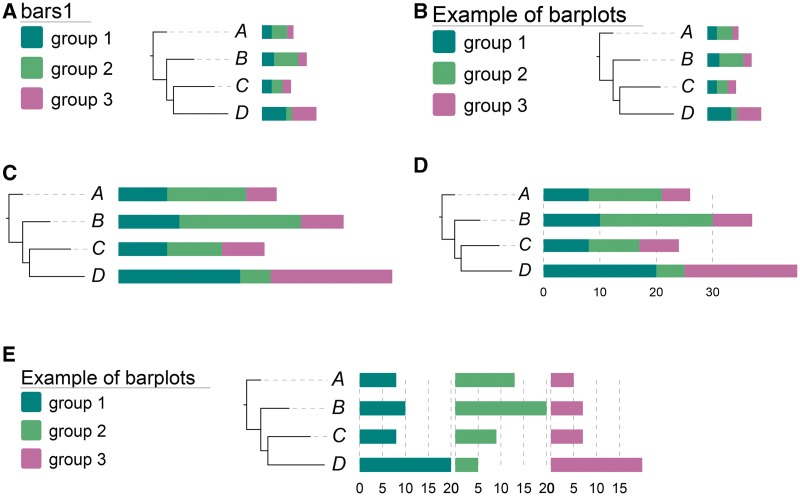
Shown in Figure 2 is a demonstration of how various aspects of the graphical annotations (bar charts in this case) can be controlled by the modifiers. The phylogenetic tree and corresponding datasets are available in Supplementary Table 2.
Figure 2.
A demonstration of how the modifiers can control various aspects of the graphical annotation; the corresponding data are available in Supplementary Table 2. (A) A simple phylogenetic tree with bar plots. By default, the title of the legend is the name of the dataset. (B) The legend title can be changed by a modifier ‘!title’. (C) The width of the graphical annotation can be specified using ‘!plotwidth’. (D) Two additional modifiers ‘!grid’ and ‘!gridlabel’ can show grid lines behind the bars and their corresponding values under the bars, respectively. (E) A simple modifier ‘!align’ can change the plot from ‘stacked’ bars to aligned-column bars. Note that the modifiers can be used in combination.
USAGE
The EvolView user interface has been designed with an emphasis on intuitive graphical elements; most of the functionalities can be accessed through their respective icons. To help users to get a quick overview of the typical workflow, we provided two step-by-step tutorials using example data. The first tutorial can be found in Supplementary Data, and the corresponding data are available in Supplementary Table 3. From this tutorial users can also learn how to prepare annotation datasets using standard spreadsheet applications such as Microsoft Excel, iWork Numbers or LibreOffice Calc. The second tutorial provides even more detailed step-by-step instruction for new users, and is available online (http://code.google.com/p/evolgenius/wiki/EvolViewQuickStart). All datasets for the two tutorials can be directly copy-pasted into our web tool. Full documentation, including detailed instructions for dataset preparation, can also be found online (http://code.google.com/p/evolgenius/wiki/EvolView).
CONCLUSION
In this article, we introduced EvolView, a comprehensive web application for visualizing, annotating and managing phylogenetic trees. Also incorporated into EvolView is an evolutionary dataset system that allows users not only to upload biologically relevant contents to be displayed graphically on the phylogenetic trees, but also to control all major aspects of the graphical annotations. To make EvolView easy to use to all the users, it comes with an intuitive and user-friendly interface that provides access to most of its functionalities through clickable icons. We believe EvolView is a useful tool to the community.
Availability and requirements
EvolView is accessible using modern browsers such as Firefox, Chrome, Opera and Safari. However, due to some compatibility issues, Internet Explorer (IE) at the moment is not supported. This website is free and open to all users and there is no login requirement. However, if users want to save their data on our server, an account is required. Registration and the account are free.
FUTURE DIRECTIONS
We aim to keep EvolView up-to-date and state-of-the-art. Future development of EvolView will include: (i) incorporation of novel data types, such as protein domains and horizontal gene transfers; (ii) incorporation of annotations on leaf as well as internal tree nodes—annotations can be entered manually or retrieved automatically from public resources; (iii) the use of faster graphical rendering engines such as HTML5 canvas to improve the drawing speed; and (iv) extending the import/export capabilities to the PhyloXML format, allowing the combined handling of phylogenetic trees and associated annotations.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–3, Supplementary Figure and Supplementary Text.
FUNDING
Funding for open access charge: Knowledge Innovation Program of the Chinese Academy of Sciences [KSCX2- EW-R-01-04].
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We are thankful to Lukas Laag and other developers for their wonderful open-source libraries used by this project; see here for a complete list (http://code.google.com/p/evolgenius/wiki/EvolViewAcknowledgements).
REFERENCES
- 1.Page RD. Visualizing phylogenetic trees using TreeView. Curr. Protoc. Bioinformatics. 2002 doi: 10.1002/0471250953.bi0602s01. Chapter 6, Unit 6 2. [DOI] [PubMed] [Google Scholar]
- 2.Huson DH, Richter DC, Rausch C, Dezulian T, Franz M, Rupp R. Dendroscope: an interactive viewer for large phylogenetic trees. BMC Bioinformatics. 2007;8:460. doi: 10.1186/1471-2105-8-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Letunic I, Bork P. Interactive tree of life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Res. 2011;39:W475–W478. doi: 10.1093/nar/gkr201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smits SA, Ouverney CC. jsPhyloSVG: a javascript library for visualizing interactive and vector-based phylogenetic trees on the web. PloS One. 2010;5:e12267. doi: 10.1371/journal.pone.0012267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jordan GE, Piel WH. PhyloWidget: web-based visualizations for the tree of life. Bioinformatics. 2008;24:1641–1642. doi: 10.1093/bioinformatics/btn235. [DOI] [PubMed] [Google Scholar]
- 6.Bingham J, Sudarsanam S. Visualizing large hierarchical clusters in hyperbolic space. Bioinformatics. 2000;16:660–661. doi: 10.1093/bioinformatics/16.7.660. [DOI] [PubMed] [Google Scholar]
- 7.Chevenet F, Brun C, Banuls AL, Jacq B, Christen R. TreeDyn: towards dynamic graphics and annotations for analyses of trees. BMC Bioinformatics. 2006;7:439. doi: 10.1186/1471-2105-7-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huerta-Cepas J, Dopazo J, Gabaldon T. ETE: a python Environment for Tree Exploration. BMC Bioinformatics. 2010;11:24. doi: 10.1186/1471-2105-11-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peterson MW, Colosimo ME. TreeViewJ: an application for viewing and analyzing phylogenetic trees. Source Code Biol. Med. 2007;2:7. doi: 10.1186/1751-0473-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pethica R, Barker G, Kovacs T, Gough J. TreeVector: scalable, interactive, phylogenetic trees for the web. PloS One. 2010;5:e8934. doi: 10.1371/journal.pone.0008934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ranwez V, Clairon N, Delsuc F, Pourali S, Auberval N, Diser S, Berry V. PhyloExplorer: a web server to validate, explore and query phylogenetic trees. BMC Evol. Biol. 2009;9:108. doi: 10.1186/1471-2148-9-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Santamaria R, Theron R. Treevolution: visual analysis of phylogenetic trees. Bioinformatics. 2009;25:1970–1971. doi: 10.1093/bioinformatics/btp333. [DOI] [PubMed] [Google Scholar]
- 13.Stover BC, Muller KF. TreeGraph 2: combining and visualizing evidence from different phylogenetic analyses. BMC Bioinformatics. 2010;11:7. doi: 10.1186/1471-2105-11-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen WH, Lercher MJ. ColorTree: a batch customization tool for phylogenic trees. BMC Res. Notes. 2009;2:155. doi: 10.1186/1756-0500-2-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chevenet F, Croce O, Hebrard M, Christen R, Berry V. ScripTree: scripting phylogenetic graphics. Bioinformatics. 2010;26:1125–1126. doi: 10.1093/bioinformatics/btq086. [DOI] [PubMed] [Google Scholar]
- 16.Paradis E, Claude J, Strimmer K. APE: analyses of Phylogenetics and Evolution in R language. Bioinformatics. 2004;20:289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- 17.Maddison DR, Swofford DL, Maddison WP. NEXUS: an extensible file format for systematic information. Syst. Biol. 1997;46:590–621. doi: 10.1093/sysbio/46.4.590. [DOI] [PubMed] [Google Scholar]
- 18.Han MV, Zmasek CM. phyloXML: XML for evolutionary biology and comparative genomics. BMC Bioinformatics. 2009;10:356. doi: 10.1186/1471-2105-10-356. [DOI] [PMC free article] [PubMed] [Google Scholar]