Abstract
LAGLIDADG homing endonucleases (LHEs) are DNA cleaving enzymes, also termed ‘meganucleases’ that are employed as gene-targeting reagents. This use of LHEs requires that their DNA specificity be altered to match sequences in genomic targets. The choice of the most appropriate LHE to target a particular gene is facilitated by the growing number of such enzymes with well-characterized activities and structures. ‘LAHEDES’ (The LAGLIDADG Homing Endonuclease Database and Engineering Server) provides both an online archive of LHEs with validated DNA cleavage specificities and DNA-binding interactions, as well as a tool for the identification of DNA sequences that might be targeted by various LHEs. Searches can be performed using four separate scoring algorithms and user-defined choices of LHE scaffolds. The webserver subsequently provides information regarding clusters of amino acids that should be interrogated during engineering and selection experiments. The webserver is fully open access and can be found at http://homingendonuclease.net.
Genome engineering and targeted gene modification is an emerging discipline in which genomes within cell lines, tissues or organisms are manipulated and altered at specified individual loci (1). Such approaches are now being used for a wide-variety of purposes, such as the correction of individual genes in patients suffering from genetic diseases (2); the targeted disruption of genes in patients afflicted by latent viral infections (3); the modification or insertion of genes in plants (4); the generation of unique transgenic stem cell lines (5); the genetic modification of animal model systems (6) and the incorporation of gene drive systems into disease vectors as part of population control strategies (7). By relying on gene-targeting enzymes to enhance the efficiency of site-specific modification, these approaches reduce the burden of intermediate screening steps and use of selectable markers that are necessary in traditional gene-targeting techniques (8–10). A variety of systems are being developed for this purpose, including site-specific recombinases (11) and transposases (12) that directly deliver gene sequences to user-defined chromosomal locations, as well as site-specific DNA cleaving enzymes. These latter systems, ranging from peptide-nucleic acid (PNA) conjugates to site-specific endonucleases, induce double strand breaks that are repaired via homologous recombination and nonhomologous end-joining, resulting in targeted gene modification and disruption, respectively (13).
Three separate protein scaffolds which generate site-specific double-stranded DNA breaks can be used for targeted gene modification: zinc-finger nucleases (ZFNs) (14), TAL effector nucleases (TALENs) (15) and LAGLIDADG homing endonucleases (LHEs; also termed ‘Meganucleases’) (16). Thus, the field of site-specific genome engineering using site-specific nucleases enjoys a wealth of structural scaffolds for the development of gene-targeting proteins. However, the structures and properties of ZFNS and TALENs differ significantly from LHEs. The first two are artificial chimeras that contain tandem arrays of modular DNA-binding domains tethered to a nonspecific nuclease domain (usually derived from the R.FokI restriction endonuclease). In contrast, LHEs are naturally occurring microbial gene-targeting proteins that are often associated with mobile introns or inteins. The DNA recognition specificities of ZFNs and TALENs are highly engineerable due to their modular architectures, but both are large proteins that must be introduced and/or expressed as two separately encoded protein chains. In contrast, LHEs are considerably more challenging to redesign, but they possess several properties that otherwise make them ideal candidates for gene targeting, including compact, stable monomeric protein folds that display exceptionally high DNA cleavage specificities (17).
The ability to routinely redirect the DNA target specificity of an LHE is therefore a highly desirable goal. A wide variety of methods have been described for this purpose, including structure-based engineering and several different selection methods [reviewed in (16)]. Until recently, only one LHE (the I-CreI endonuclease) had been successfully engineered and used for the targeted modification of actual physiological coding sequences (18,19), while several additional LHEs (including I-MsoI, I-SceI and I-AniI) had been redesigned in a more limited manner [also reviewed in (16)].
A recent bioinformatics analysis (20) demonstrated that (i) many hundreds of recognizable LHE genes can be found within microbial genomic sequence databases; (ii) a significant fraction of those genes encode active endonucleases; (iii) their target sites can often be determined by a combination of comparative genomic analyses and biochemical experimentation; and (iv) even closely related LHE homologs can display considerably diverged DNA cleavage specificities. These observations were exploited to rapidly generate a novel gene-targeting protein that was highly active against the endogenous human monoamine oxidase B gene (20). Based on those observations, it seems clear that the natural diversity of LHEs can be further exploited in the field of gene targeting, by providing a wealth of initial scaffolds that collectively would reduce the challenge of engineering reagents for individual gene targets. This concept was also suggested in an earlier analysis of putative homing endonuclease genes, which was subsequently organized into the HomeBase webserver (http://homebase-search.tau.ac.il) (21).
The identification and characterization of active LHEs has motivated the development of a dedicated LHE database and webserver, described below, that could serve both as an archive of those LHEs that have been sufficiently characterized to be used as starting points for creating gene-targeting proteins, and as a portal for identifying combinations of endonuclease scaffolds and DNA targets that could be used for targeted gene modification. The resource is not an all-inclusive database of LHEs [of which there are hundreds that are known, most of which are already collected and cataloged in InBase (22), ReBase (23), GISSD (24) and HomeBase (21)] but rather a resource to facilitate the creation of gene-targeting proteins from those LHES that meet at least the three following criteria:
The exact boundaries and center of their DNA target sites are known;
The orientation and positioning of their N- and C-terminal enzyme domains, relative to the two halves of the DNA target site are known; and
The identity of amino acid residues that are located within close proximity to clusters of nucleotide bases in the DNA target site are known.
This online resource consists of five tools that are accessible from a top-level group of menu items: an Endonuclease Browser that provides the identity and basic properties of well-characterized LHEs and their target sites, Entry tools for adding new endonucleases that meet the criteria listed above, their validated target sites and several additional properties, Genomic Search tools for the identification of potential target sites that might be modified through the use of wild-type or engineered LHEs, and Additional Information menus that provide additional background information both on homing endonuclease biology and on the use of the webserver. The entire resource is designed to be extremely simple and intuitive to use, and instructions for all key steps are provided as online popups that are accessed directly at the individual points of operation for all key functions.
LHE DATABASE
Enzyme and target site entry
The first core functionality of the LAGLIDADG Homing Endonuclease Database and Engineering Server (LAHEDES) resource is an updateable collection of wild-type and engineered LHEs that have been shown to cleave precisely defined DNA sequences corresponding to their target sites in original host gene, and for which the exact positioning and orientation of the target site relative to the LHE’s N- and C-terminal domains are known. In the database, the target sites are entered and listed as 22-bp sequences that exactly span the center of four consecutive bases (termed the ‘central four target region’) that are converted into mutually cohesive four-base 3′-overhangs by all known LHEs. The orientation of the target site relative to the bound endonuclease (i.e. which half-sites are engaged by the N- and C-terminal domains of the LHE, respectively) is defined based on biochemical and/or structural analyses. In this convention, the left half-site (basepairs −11 to −1) is contacted by residues from the N-terminal LHE domain, while the right half-site (basepairs +1 to +11) is bound by the C-terminal domain.
The ‘Entry’ tool for addition of new LHEs to the database provides individual fields for the enzyme name, its amino acid sequence (entered as a FASTA character string), its DNA target site (entered from basepair −11 to +11 as described above) and a notes field for the entry of additional information for the endonuclease. This information usually includes (i) the host organism and corresponding host gene; (ii) the relationship of the LHE to any surrounding self-splicing elements (introns or inteins) or its existence as a ‘stand-alone’ endonuclease; (iii) links to genomic or structural information for the endonuclease; and (iv) the nature of experiments that have validated the LHE's activity.
In addition to wild-type LHEs, entry fields are available to archive ‘chimeric’ LHEs (which correspond to various types of structural fusions between N- and C-terminal LHE domains) (25–27) ‘monomerized’ LHEs (which correspond to homodimeric endonucleases that have been converted into single-chain reagents through the fusion and tethering of two protein subunits with an artificial peptide linker) (28,29) and fully redesigned LHEs (for those that have been entirely retargeted to genomic target sites and that have been biochemically characterized at a level that matches their wild-type parental enzyme) (20). In each case, the same general entry tools and note fields are provided, with individualized instructions for the information required for adequate archiving. Finally, it is also possible to enter the names, protein sequences and biological origins of ‘pseudo-endonucleases’, which correspond to LHE genes that do not appear to encode stable or active enzymes (a category motivated by the observation that a significant fraction of homing endonuclease genes have accumulated disabling mutations subsequent to the successful invasion of their host gene) (30).
The rules for naming new LHEs, following the conventions described in Roberts et al. (31) are explicitly described and followed by the LAHEDES database, including instructions to examine the REBASE database (23) to ensure that unique acronyms for novel host species and genus are chosen. In general, the entry tool recommends adding an additional character to the end of the acronym to further denote the source of the LHE in cases where a newly discovered enzyme is derived from a mitochondrial (‘M’) or chloroplast (‘C’) genome. Certain enzymes that have already enjoyed a long history in the published literature and corresponding databases (such as I-SceI, I-CreI, etc.) maintain their historical names.
As mentioned above, the browser is limited to those LHEs that have been sufficiently characterized that they can be utilized for the creation of gene-targeting reagents (a list that currently spans 24 enzymes). However, the resource is open-ended and available for deposition of any LHEs that meet the necessary criterion for protein engineering and selection experiments.
Specificity profile (position weight matrix) entry
In addition to the exact sequence of their naturally occurring DNA targets (which usually corresponds to the site in the host gene which is cleaved by the homing endonuclease, resulting in invasion of that site by the mobile HE gene and surrounding DNA sequences), the specificity of a given LHE in many cases is also represented in the database by an additional Position Weight Matrix (PWM) (32). This PWM takes into account any positions in the DNA target where base substitutions are tolerated in cleavage experiments (in other words, positions in the DNA target that are recognized with reduced fidelity). This feature of LHEs (the ability to tolerate individual polymorphisms in their natural target sites) is an evolutionary consequence of the need to accommodate the natural drift in the sequence of their host gene targets, and to effectively execute ectopic transfers into new host genes that contain related target sites when the opportunity arises (30,33). In particular, LHE reading frames that are found within protein-coding genes often tend to display reduced fidelity at positions corresponding to ‘wobble’ positions in the host gene reading frame (33) [a feature of DNA recognition that is also explicitly accounted for in the HomeBase webserver (21)]. The use of an experimentally determined PWM as a search-scoring matrix exploits this property, but reduces the risk of false positives that might arise from enzymes that depart from this general rule for recognition specificity.
The need to account for relaxed fidelity in LHE–DNA recognition is important in enzyme redesign experiments, where mismatches between the wild-type target site and the desired DNA target which correspond to well-tolerated polymorphisms can be accommodated with minimal effort during the engineering or selection process. Accounting for the specificity profile of an LHE is also important in predicting potential genomic ‘off-target’ sites, as well as regions of the protein–DNA interface where specificity may be enhanced through additional engineering.
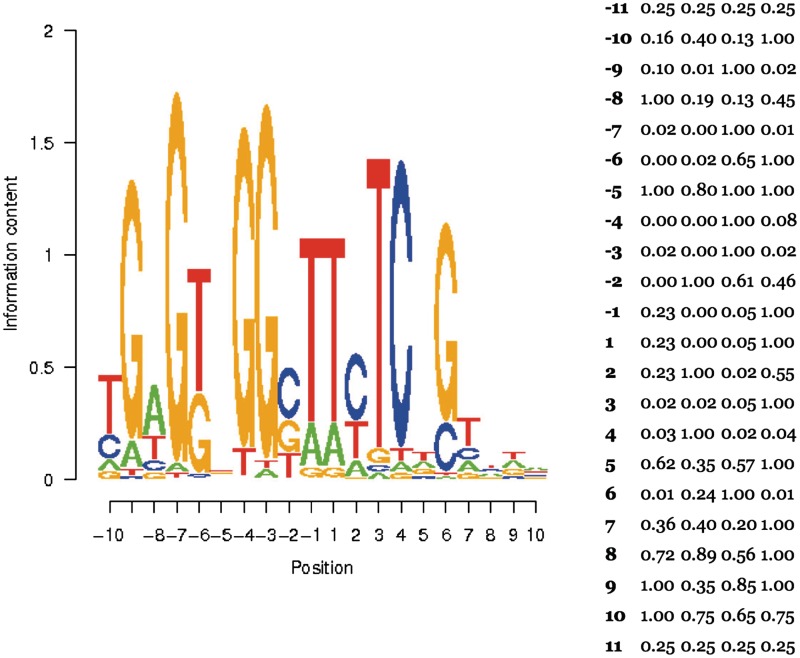
PWMs are entered into the database with a separate entry tool, using information derived either from direct biochemical measurements of the enzyme's ability to cleave alternative target site variants (34) or from experiments that identify collections of cleavable DNA target variants from pools of partially randomized sites (33). In either case, the relative ability of the enzyme to accommodate each possible base at each possible position in the target site is usually entered using a scale of 1.0 (wild-type cleavage activity or recovery for a given base) to 0.0 (complete lack of cleavage activity or recovery for a given base). The resulting PWM values are displayed by the database both as a sequence logo plot, and as raw text that can be incorporated into separate computational analyses (Figure 1).
Figure 1.
The specificity profile (PWM) for the I-AniI LHE. Specificity profiles, reported as PWMs that have been experimentally determined for a number of LHEs can be entered individually and are available on the site as part of the browser functionality. These matrices are displayed in graphical format as a sequence logo plot (left) and in tabular format. The ability of the enzyme to cleave DNA targets that contain a given nucleotide at a specific position within the target site is indicated by letter height, and positions with greater overall information content are more specifically recognized. The profile displayed in this figure was determined in a series of biochemical experiments where the cleavability of target site variants was individually measured (34).
DNA contact module and redesign mutation entry
In addition to target sites and PWMs, the database accommodates lists of residues for each homing endonuclease that might be potentially useful in any redesign or selection process, and links the information within those lists to the output of target site searches (described below). After entry of a new wild-type LHE, the entry tool for ‘contact modules’ is then populated by that enzyme in a drop-down menu that allows an investigator to list those amino acids that the investigator believes to be within contact distance to individual clusters of target sites basepairs. This entry tool is arranged for each enzyme into overlapping three base ‘modules’ across the target site, that can be incorporated into experiments to alter the enzyme's recognition specificity within that same DNA module. For example, the I-OnuI endonuclease displays direct and water-mediated contacts between the −11 to −9 bp positions and at least seven separate amino acid side-chains: Asn 32, Lys 34, Ser 35, Ser 36, Val 37, Gly 38 and Ser 40 (20); all are listed as residues that comprise the ‘DNA contact module’ for those three corresponding basepairs.
The choice of amino acids that should populate individual DNA contact modules can be derived either from crystallographic structures of a corresponding LHE–DNA complex, or from homology models constructed from such available structures. The contact residues entered into the database for the I-CreI homodimeric LHE and the I-OnuI monomeric LHE have been extensively validated for engineering purposes, and those can be used as an initial reference for the entry of corresponding residue lists for homologous LHEs. The premise behind entering amino acid contact lists in a modular format is based on past experimental observations that sequence recognition within the LHE–DNA interface often displays considerable context dependence and cross-talk between adjacent positions (28,34,35), such that even when only 1-bp substitution is required in the novel target site, best practice often dictates that residues involved in contacts to the ‘n − 1’ and ‘n + 1’ basepairs should also be subjected to redesign or selection.
For many enzymes in the database, specific mutations in the DNA contacting residues described above have been shown to be associated with defined alterations in target sequence specificity (20,34,36). For example, incorporation of three amino acid substitutions in the monomerized version of I-MsoI (I30E, S43R andI85Y) results in a shift in specificity from a wild-type A:T basepair at position −8 (termed ‘−8A’ in the database) to preferential recognition of a G:C basepair at the same position (‘−8G’) (35). In addition to the entry tool for wild-type DNA contacting residues, an additional ‘Specificity Changing Mutation Entry’ tool is provided that allows the archiving of such substitutions in the protein scaffold and the corresponding change in DNA base preference in the altered target site. Similar to the list of wild-type DNA contacting residues, the identities of such mutations are provided to the user both in the endonuclease browser tool, and as a subsequent link from the output of the genomic sequence target site searches to aid in the design of engineering or selection experiments.
LHE SEARCH AND ENGINEERING SERVER
The second core functionality of the LAHEDES resource is the ability to search individual genes or collections of genes for potential matches against LHE target sites based on four separate search criteria:
Searches for DNA sequences that exhibit identity across the central 4 bp of an LHE target site, because those bases are usually not in direct contact with amino acid side chains. Therefore, specificity changes at these basepairs are often not easily achieved through protein engineering or selection (34). Additional experiments on individual LHE enzymes that further profile the ability of the enzyme to accommodate individual basepair substitutions in the ‘central four’ region of the target site expands the number of hits within a genomic query. For those enzymes where such information is available, the ‘Central Four’ search algorithm takes such polymorphisms into account when returning potential targets and allows such sites to appear on the list of target site hits.
Searches for highest possible sequence identity (fewest DNA basepair mismatches) across a 22-bp DNA sequence in the genomic query relative to an LHE’s entire physiological target site. Some redesign experiments indicate, not surprisingly, that the most effective way to maintain wild-type levels of affinity, activity and overall specificity is to maintain the highest degree of sequence similarity to the wild-type target (20).
For those LHEs that have had their complete specificity profile determined, the user is provided with the opportunity to search and score potential targets using a ‘Specificity Profile PWM’ described above. In this search algorithm, the penalty for a mismatch between a protein’s natural target site and a potential genomic target is differentially weighted depending on the fidelity of recognition displayed at each DNA basepair position. The difference between the two search strategies described in (ii) versus (iii) is simple: the use of a simple identity matrix will return those potential targets with the fewest mismatches relative to the enzyme's wild-type recognition site, while the use of a scoring matrix that accounts for recognition degeneracy would indicate sites that might be more distantly related to the protein's wild-type recognition site, while nevertheless indicating more tractable gene-targeting sites.
The I-OnuI LHE has been systematically tested for its ability to be retargeted towards each possible altered base triplet across the entire length of the DNA target site, using a high throughput method that combines yeast surface display and flow cytometry (37) (J. Jarjour, unpublished data). For that enzyme, genomic searches are therefore possible using corresponding modular scoring matrices, returning lists of potential target sites to the investigator that might be accessible for modification through a subsequent experimental strategy that makes use of highthroughput selection from pools of enzyme variants that are randomized at corresponding DNA-contacting residues.
The search algorithm also allows the user to search for genomic sequences that might be targeted through the application of validated ‘chimeric’ homing endonucleases (where N- and C-terminal domains or DNA contacting surfaces of unrelated LHEs have been fused or individually modified to create scaffolds that can recognize corresponding chimeric DNA target sites) (25–27).
Following genomic target site searches using any of the four methods outlined above (along with user-chosen endonucleases for the search that can span all or a smaller subset of those LHEs that are available in the database), a top-scoring list of putative target sequences is returned in a graphical user interface, alongside the enzyme's wild-type target for comparison (Figure 2a). The output, which can be saved to a text file, indicates the position and identity of all basepairs in the potential target that differs from the enzyme's wild-type recognition site, and provides links from each base in the candidate targets to information that has been entered about amino acids and mutations in the LHE that might influence specificity and redesign at that position.
Figure 2.
Genomic search results. The human monoamine oxidase B (MAOB) gene was searched for potential target sites matches using the I-OnuI LHE and either the ‘Identity’ algorithm (a) or the ‘Modular Engineerability’ algorithm (b). For both searches, a list of best matches is shown, together with their position in the query sequence, orientation (forward or reverse) and scores. The top scoring hit in the ‘Identity’ Search (Panel A, top MAOB sequence) was targeted for cleavage at the endogenous chromosomal MAOB locus by an engineered version of I-OnuI (20) (listed as ‘I-OnuI(E2)’ in the endonuclease browser, see Figure 1). Throughout the target site hits, mismatches are shown as lower base bases. In the output from a Module Search, each score is linked to a more detailed view (c) illustrating how well each DNA module scores using an ‘engineerability’ matrix for I-OnuI. Modules are represented by individual bars; those that score favorably in a given target are colored blue, while those that do not score as favorably are colored red. Nucleotides that directly match the wild type target are indicated by vertical bars. The engineerability scoring scale (0 to 10; 10 is best) is shown to the right. Clicking on individual bases or modular bars in the search output provides links to lists of amino acids (d) and (if they are available) mutations in the protein that are associated with recognition at those positions in the target.
While the search output using the first three scoring algorithms (‘Central 4 Match’, ‘Target Identity’ and ‘PWM Search’) is arranged to clearly display the conservation or mismatch at each individual DNA basepair, the output for the ‘Modular’ search displays individual genomic sequences in the form of information regarding the ‘engineerability’ of each protein–DNA module that harbors one or more basepair changes (Figure 2b). As described above, the individual regions and basepairs in potential target sites are linked to previously entered information regarding the identity of amino acids and the corresponding mutations that are involved in the recognition of those positions.
CONCLUSIONS
The field of genome and gene engineering is currently experiencing a rapid increase in the amount of information and data relating to the creation of gene-targeting proteins derived from the LHE family. These include: (i) the continuing identification of new LHEs from worldwide microbial and metagenomic sequencing projects and (ii) the generation of a large numbers of engineered LHE variants with altered DNA recognition properties. One manner in which the full potential and impact of this information can be fully exploited is through the development and maintenance of a computational database and search engine for LHE proteins.
Although LHEs have historically been considerably more difficult to engineer for altered DNA recognition and cleavage properties relative to their artificial, modular ZFN and TALEN counterparts, there still exists considerable industrial and academic activity and motivation to reduce or eliminate the technical gulf that separates them from routine application in genome engineering applications. It is likely that the combination of highthroughput modular screens for altered specificity and the increasing numbers of wild-type and chimeric LHE protein scaffolds, along with the ability to apply deep sequencing methods to resulting pools of highly active enzyme variants, may soon drive a significant reduction in the time and cost of assembling artificial LHEs for the life science community.
FUNDING
Fred Hutchinson Cancer Research Center and NIH [RL1 CA133833]. Funding for open access charge: National Institutes of Health [RL1 CA133833].
Conflict of interest statement. Barry Stoddard is a Founder, and Jordan Jarjour is an employee, of Pregenen Inc., which is focused on the development and engineering of LHEs for gene targeting applications.
REFERENCES
- 1.McMahon MA, Rahdar M, Porteus M. Gene editing: not just for translation anymore. Nat. Methods. 2012;9:28–31. doi: 10.1038/nmeth.1811. [DOI] [PubMed] [Google Scholar]
- 2.Ellis BL, Hirsch ML, Porter SN, Samulski RJ, Porteus MH. Zinc-finger nuclease-mediated gene correction using single AAV vector transduction and enhancement by Food and Drug Administration-approved drugs. Gene Ther. 2012 doi: 10.1038/gt.2011.211. January 19 (doi:10.1038/gt.2011.211; epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cannon P, June C. Chemokine receptor 5 knockout strategies. Curr. Opin. HIV AIDS. 2011;6:74–79. doi: 10.1097/COH.0b013e32834122d7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim S, Kim JS. Targeted genome engineering via zinc finger nucleases. Plant Biotechnol. Rep. 2011;5:9–17. doi: 10.1007/s11816-010-0161-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Osiak A, Radecke F, Guhl E, Radecke S, Dannemann N, Lutge F, Glage S, Rudolph C, Cantz T, Schwarz K, et al. Selection-independent generation of gene knockout mouse embryonic stem cells using zinc-finger nucleases. PloS One. 2011;6:e28911. doi: 10.1371/journal.pone.0028911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ekker SC. Zinc finger-based knockout punches for zebrafish genes. Zebrafish. 2008;5:121–123. doi: 10.1089/zeb.2008.9988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Windbichler N, Menichelli M, Papathanos PA, Thyme SB, Li H, Ulge UY, Hovde BT, Baker D, Monnat RJ, Jr, Burt A, et al. A synthetic homing endonuclease-based gene drive system in the human malaria mosquito. Nature. 2011;473:212–215. doi: 10.1038/nature09937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Glaser S, Anastassiadis K, Stewart AF. Current issues in mouse genome engineering. Nat. Genet. 2005;37:1187–1193. doi: 10.1038/ng1668. [DOI] [PubMed] [Google Scholar]
- 9.Tzfira T, White C. Towards targeted mutagenesis and gene replacement in plants. Trends Biotechnol. 2005;23:567–569. doi: 10.1016/j.tibtech.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 10.Posfai G, Kolisnychenko V, Bereczki Z, Blattner FR. Markerless gene replacement in Escherichia coli stimulated by a double-strand break in the chromosome. Nucleic Acids Res. 1999;27:4409–4415. doi: 10.1093/nar/27.22.4409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gersbach CA, Gaj T, Gordley RM, Mercer AC, Barbas CF., 3rd Targeted plasmid integration into the human genome by an engineered zinc-finger recombinase. Nucleic Acids Res. 2011;39:7868–7878. doi: 10.1093/nar/gkr421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kettlun C, Galvan DL, George AL, Jr, Kaja A, Wilson MH. Manipulating piggyBac transposon chromosomal integration site selection in human cells. Mol. Ther. J. Am. Soc. Gene Ther. 2011;19:1636–1644. doi: 10.1038/mt.2011.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aiba Y, Sumaoka J, Komiyama M. Artificial DNA cutters for DNA manipulation and genome engineering. Chem. Soc. Rev. 2011;40:5657–5668. doi: 10.1039/c1cs15039a. [DOI] [PubMed] [Google Scholar]
- 14.Carroll D. Genome engineering with zinc-finger nucleases. Genetics. 2011;188:773–782. doi: 10.1534/genetics.111.131433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bogdanove AJ, Voytas DF. TAL effectors: customizable proteins for DNA targeting. Science. 2011;333:1843–1846. doi: 10.1126/science.1204094. [DOI] [PubMed] [Google Scholar]
- 16.Stoddard BL. Homing endonucleases: from microbial genetic invaders to reagents for targeted DNA modification. Structure. 2011;19:7–15. doi: 10.1016/j.str.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paques F, Duchateau P. Meganucleases and DNA double-strand break-induced recombination: perspectives for gene therapy. Curr. Gene Ther. 2007;7:49–66. doi: 10.2174/156652307779940216. [DOI] [PubMed] [Google Scholar]
- 18.Gao H, Smith J, Yang M, Jones S, Djukanovic V, Nicholson MG, West A, Bidney D, Falco SC, Jantz D, et al. Heritable targeted mutagenesis in maize using a designed endonuclease. Plant J Cell Mol. Biol. 2010;61:176–187. doi: 10.1111/j.1365-313X.2009.04041.x. [DOI] [PubMed] [Google Scholar]
- 19.Munoz IG, Prieto J, Subramanian S, Coloma J, Redondo P, Villate M, Merino N, Marenchino M, D'Abramo M, Gervasio FL, et al. Molecular basis of engineered meganuclease targeting of the endogenous human RAG1 locus. Nucleic Acids Res. 2011;39:729–743. doi: 10.1093/nar/gkq801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takeuchi R, Lambert AR, Mak AN, Jacoby K, Dickson RJ, Gloor GB, Scharenberg AM, Edgell DR, Stoddard BL. Tapping natural reservoirs of homing endonucleases for targeted gene modification. Proc. Natl Acad. Sci. USA. 2011;108:13077–13082. doi: 10.1073/pnas.1107719108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barzel A, Privman E, Peeri M, Naor A, Shachar E, Burstein D, Lazary R, Gophna U, Pupko T, Kupiec M. Native homing endonucleases can target conserved genes in humans and in animal models. Nucleic Acids Res. 2011;39:6646–6659. doi: 10.1093/nar/gkr242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perler FB. InBase: the Intein Database. Nucleic Acids Res. 2002;30:383–384. doi: 10.1093/nar/30.1.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roberts RJ, Vincze T, Posfai J, Macelis D. REBASE–a database for DNA restriction and modification: enzymes, genes and genomes. Nucleic Acids Res. 2010;38:D234–D236. doi: 10.1093/nar/gkp874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou Y, Lu C, Wu QJ, Wang Y, Sun ZT, Deng JC, Zhang Y. GISSD: Group I Intron Sequence and Structure Database. Nucleic Acids Res. 2008;36:D31–D37. doi: 10.1093/nar/gkm766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chevalier BS, Kortemme T, Chadsey MS, Baker D, Monnat RJ, Stoddard BL. Design, activity, and structure of a highly specific artificial endonuclease. Mol. Cell. 2002;10:895–905. doi: 10.1016/s1097-2765(02)00690-1. [DOI] [PubMed] [Google Scholar]
- 26.Grizot S, Epinat JC, Thomas S, Duclert A, Rolland S, Paques F, Duchateau P. Generation of redesigned homing endonucleases comprising DNA-binding domains derived from two different scaffolds. Nucleic Acids Res. 2010;38:2006–2018. doi: 10.1093/nar/gkp1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Silva GH, Belfort M, Wende W, Pingoud A. From monomeric to homodimeric endonucleases and back: engineering novel specificity of LAGLIDADG enzymes. J. Mol. Biol. 2006;361:744–754. doi: 10.1016/j.jmb.2006.06.063. [DOI] [PubMed] [Google Scholar]
- 28.Grizot S, Smith J, Daboussi F, Prieto J, Redondo P, Merino N, Villate M, Thomas S, Lemaire L, Montoya G, et al. Efficient targeting of a SCID gene by an engineered single-chain homing endonuclease. Nucleic Acids Res. 2009;37:5405–5419. doi: 10.1093/nar/gkp548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li H, Pellenz S, Ulge U, Stoddard BL, Monnat RJ., Jr Generation of single-chain LAGLIDADG homing endonucleases from native homodimeric precursor proteins. Nucleic Acids Res. 2009;37:1650–1662. doi: 10.1093/nar/gkp004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Posey KL, Koufopanou V, Burt A, Gimble FS. Evolution of divergent DNA recognition specificities in VDE homing endonucleases from two yeast species. Nucleic Acids Res. 2004;32:3947–3956. doi: 10.1093/nar/gkh734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roberts RJ, Belfort M, Bestor T, Bhagwat AS, Bickle TA, Bitinaite J, Blumenthal RM, Degtyarev S, Dryden DT, Dybvig K, et al. A nomenclature for restriction enzymes, DNA methyltransferases, homing endonucleases and their genes. Nucleic Acids Res. 2003;31:1805–1812. doi: 10.1093/nar/gkg274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Henikoff S. Scores for sequence searches and alignments. Curr. Opin. Struct. Biol. 1996;6:353–360. doi: 10.1016/s0959-440x(96)80055-8. [DOI] [PubMed] [Google Scholar]
- 33.Scalley-Kim M, McConnell-Smith A, Stoddard BL. Coevolution of a homing endonuclease and its host target sequence. J. Mol. Biol. 2007;372:1305–1319. doi: 10.1016/j.jmb.2007.07.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thyme SB, Jarjour J, Takeuchi R, Havranek JJ, Ashworth J, Scharenberg AM, Stoddard BL, Baker D. Exploitation of binding energy for catalysis and design. Nature. 2009;461:1300–1304. doi: 10.1038/nature08508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ashworth J, Taylor GK, Havranek JJ, Quadri SA, Stoddard BL, Baker D. Computational reprogramming of homing endonuclease specificity at multiple adjacent base pairs. Nucleic Acids Res. 2010;38:5601–5608. doi: 10.1093/nar/gkq283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ashworth J, Havranek JJ, Duarte CM, Sussman D, Monnat RJ, Jr, Stoddard BL, Baker D. Computational redesign of endonuclease DNA binding and cleavage specificity. Nature. 2006;441:656–659. doi: 10.1038/nature04818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jarjour J, West-Foyle H, Certo MT, Hubert CG, Doyle L, Getz MM, Stoddard BL, Scharenberg AM. High-resolution profiling of homing endonuclease binding and catalytic specificity using yeast surface display. Nucleic Acids Res. 2009;37:6871–6880. doi: 10.1093/nar/gkp726. [DOI] [PMC free article] [PubMed] [Google Scholar]