Abstract
The enrichment analysis is a standard procedure to interpret ‘omics’ experiments that generate large gene lists as outputs, such as transcriptomics and protemics. However, despite the huge success of enrichment analysis in these classes of experiments, there is a surprising lack of application of this methodology to survey other categories of large-scale biological data available. Here, we report Kegg Orthology enrichMent-Online DetectiOn (KOMODO), a web tool to systematically investigate groups of monophyletic genomes in order to detect significantly enriched groups of homologous genes in one taxon when compared with another. The results are displayed in their proper biochemical roles in a visual, explorative way, allowing users to easily formulate and investigate biological hypotheses regarding the taxonomical distribution of genomic elements. We validated KOMODO by analyzing portions of central carbon metabolism in two taxa extensively studied regarding their carbon metabolism profile (Enterobacteriaceae family and Lactobacillales order). Most enzymatic activities significantly biased were related to known key metabolic traits in these taxa, such as the distinct fates of pyruvate (the known tendency of lactate production in Lactobacillales and its complete oxidation in Enterobacteriaceae), demonstrating that KOMODO could detect biologically meaningful differences in the frequencies of shared genomic elements among taxa. KOMODO is freely available at http://komodotool.org.
INTRODUCTION
High-throughput experiments are now a standard procedure in the quest for understanding intrinsically complex biological systems. A widely adopted approach to help scientists to interpret the wealth of data produced by such ‘omics’ experiments is the gene set enrichment analysis (1). The archetypal use of enrichment analysis thus far has been the search for significantly biased distribution of biological roles in ‘omics’ experiments that generate large gene lists as final result, such as transcriptomics and proteomics (2). Despite the considerable success of enrichment analysis when used to analyze these classes of experiments, there are relatively few applications of this strategy to survey other categories of ‘omics’ data available. Although some interesting approaches have been developed, such as (i) comparison of individual genomes in the Gene Ontology space (3); (ii) analysis of phenotypic data (4); and (iii) study of promoter regions associated to distinct gene sets (5); there is a vast unexplored array of possibilities concerning the use of enrichment analysis to evaluate other biological systems besides those surveyed by transcriptomics and proteomics experiments.
In this study, we report the development of a web tool (KOMODO, Kegg Orthology enrichMent-Online DetectiOn) that uses an enrichment analysis strategy to detect biased frequencies in groups of homologous genes when comparing an especially important biological entity: the monophyletic taxon. To the best of our knowledge, this is the first time that this entity is being contemplated with such enrichment approach. Additionally, KOMODO goes beyond simply listing the homologous groups differentially distributed in the distinct taxa analyzed: they are also displayed in their proper biochemical pathways, allowing users to quickly query their own taxa and pathways of interest in a visual and explorative way. To validate our methodology, we analyzed the frequencies of homologous groups in portions of the central carbon metabolism (glycolysis and citrate cycle), comparing two taxa that are extensively genetically and phenotypically characterized regarding their carbon metabolism capabilities (Enterobacteriaceae family and Lactobacillales order). Most of the enzymatic activities differentially represented were directly related to already known key metabolical traits in these taxa. The results found for these extremely well-characterized taxa suggest that enrichment analysis is a tool capable of detecting biologically meaningful biases in the distribution of shared genomic elements across taxa, and inaugurates the use of enrichment analysis as a tool to systematically characterize the taxon under the ‘omics’ paradigm. KOMODO is freely available for use at http://komodotool.org.
MATERIALS AND METHODS
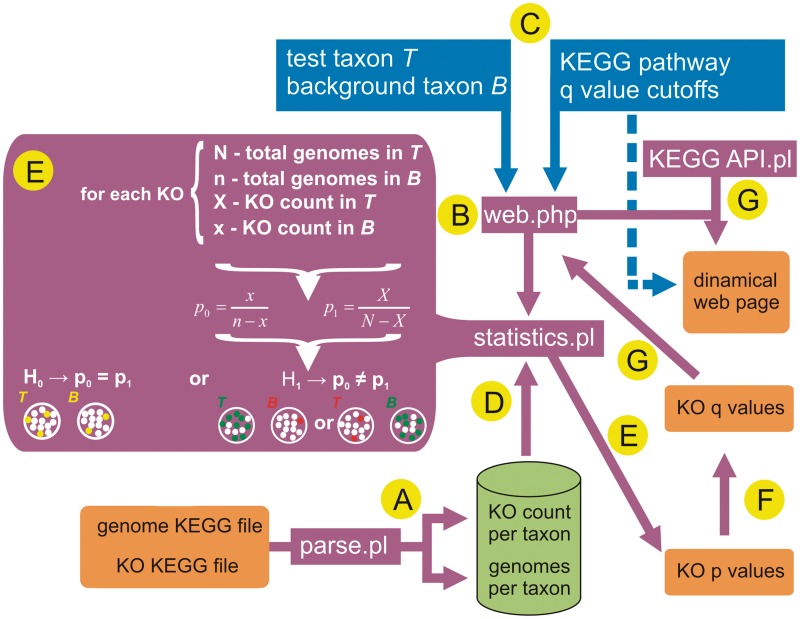
The main steps for the development of KOMODO are represented in Figure 1. A deeper explanation of this process is described in the Supplementary File 1. To develop KOMODO we initially downloaded, parsed and stored genomic data from KEGG (6) in a local database that contains, for virtually all taxa present in the KEGG database, their total number of genomes in KEGG and their number of genomes that possess a given KEGG Orthology (KO) group—the KEGG database object to represent sets of homologous genes (Figure 1, step A). For any analysis to be performed (Figure 1, step B), the user is required to input three parameters: the two taxa being compared (a test taxon T and a background taxon B), a KEGG pathway for visualization, and two values for upper and lower significance cut-offs (Figure 1, step C, blue squares). With these parameters KOMODO queries the local database to obtain four numbers for each KO: N (number of genomes in T), n (number of genomes in B), X (number of genomes in T with the specified KO) and x (number of genomes in B with the specified KO; Figure 1, step D). After this step, KOMODO performs a chi-square test to evaluate if the ratios p0 (x/(t − x)) and p1 (X/(T − X)) derivate from the same distribution by testing the likeliness of null (H0, p0 = p1) and alternative (H1, p0 ≠ p1) hypotheses (Figure 1, step E). The widely adopted false discovery rate (FDR) is used to control Type 1 error in this multiple hypothesis testing scenario (Figure 1, step F). The results found are displayed in two manners (Figure 1, step G). KOMODO generates a clickable webpage through the KEGG API tool to allow users to explore the KO distribution in a particular biochemical pathway. Specifically, under- or over-represented enzymatic activities are displayed in shades of red and green, respectively, colored according to the q-values found. It is also possible to download the entire results found as a text file for use in further analyses. KOMODO takes roughly a minute or less to generate the results.
Figure 1.
Pipeline used for implementation of KOMODO. Orange boxes denote flat files from KEGG or generated by KOMODO. Purple boxes represent programs developed in this study. Green cylinder represents the relational database generated for this study. Blue boxes indicate the user-supplied information needed for KO enrichment analysis. The sequential steps for analysis are highlighted by yellow dots. (A) Parsing of KEGG information to generate KO count and genome number per taxa; data stored in a local relational database. (B) Program ‘web.php’ coordinates the entire pipeline by getting user-defined parameters (step C) to search in the relational database (step D), make the statistical testing (steps E, F and G), and generate a dynamic webpage and a flat file as final results (step G).
RESULTS AND DISCUSSION
To evaluate the capacity of KOMODO to detect biologically meaningful differences in the frequencies of groups of homologous genes in distinct taxa, we studied the distribution of enzymatic activities from two central carbon pathways (glycolysis and citrate cycle) in two of the most phenotypically well-characterized bacterial taxa regarding carbon metabolism: (i) the Enterobacteriaceae family (from now on referred to as the ENT group) and (ii) the Lactobacillales order (from now on referred to as the LAB group). The ENT group is composed of Gram-negative, LPS producing, facultative anaerobic bacilli able to completely oxidize a variety of sugars to CO2 when in aerobic growth through glycolysis, citrate cycle and oxidative phosphorylation or to several other end products (such as lactate and ethanol) through fermentative processes when in anaerobic growth (7). The LAB group is composed of Gram-positive, anaerobic or microaerophiles bacilli, able to metabolize several sugars through glycolysis to produce lactate, a key marker of this taxon, as well as other end products, such as formate, ethanol and acetate (8). Some LAB members can also use pyruvate to fuel an incomplete version of the citrate cycle to generate intermediates for the synthesis of amino acids (8,9).
The ENT and LAB groups are expected to have common enzymatic capabilities due to the shared portion of their evolutionary history as well as to possible convergent events caused by their occurrence in similar metabolic niches (sugar-rich environments), but they also possess known key lineage-specific metabolic traits, such as lactate production in LAB or the capability of growth in a medium containing citrate as single carbon source in ENT. Therefore, they constitute two reliable gold-standard taxa to evaluate the ability of KOMODO to highlight their known shared and specific metabolic traits. We chose to analyze the ENT and LAB groups by the indirect comparison of them using as background group the most specific taxon to encompass them (the Bacteria Domain). A direct comparison of the test taxa against each other can be found in the Supplementary File 1. The q-value cut-offs for significance were defined as 0.01 (cut-off 1) and 0.00001 (cut-off 2). The test taxa ENT and LAB and the background taxon Bacteria contained 107, 74 and 1005 complete genomes, respectively. The main findings of this analysis follow below, but we strongly encourage readers to examine the Supplementary File 1 for a thorough description of our findings.
Glycolysis
Glycolysis is the common name of several partially overlapped biochemical pathways that anaerobically convert glucose to a key intermediate of metabolism, pyruvate, producing ATP and NADH during the process (10). After pyruvate production, several metabolic branches are known, such as the several classes of fermentative processes (alcoholic, lactic, malonic, etc) that restore the oxidative balance of the cell. Another alternative at this point is to further oxidize pyruvate through the citric acid cycle and the oxidative phosphorylation, also restoring the oxidative balance and producing considerable quantities of NADH and ATP (11). The Figures 2 and 3 depict the analysis of the glycolytic pathway for the ENT and LAB groups, respectively.
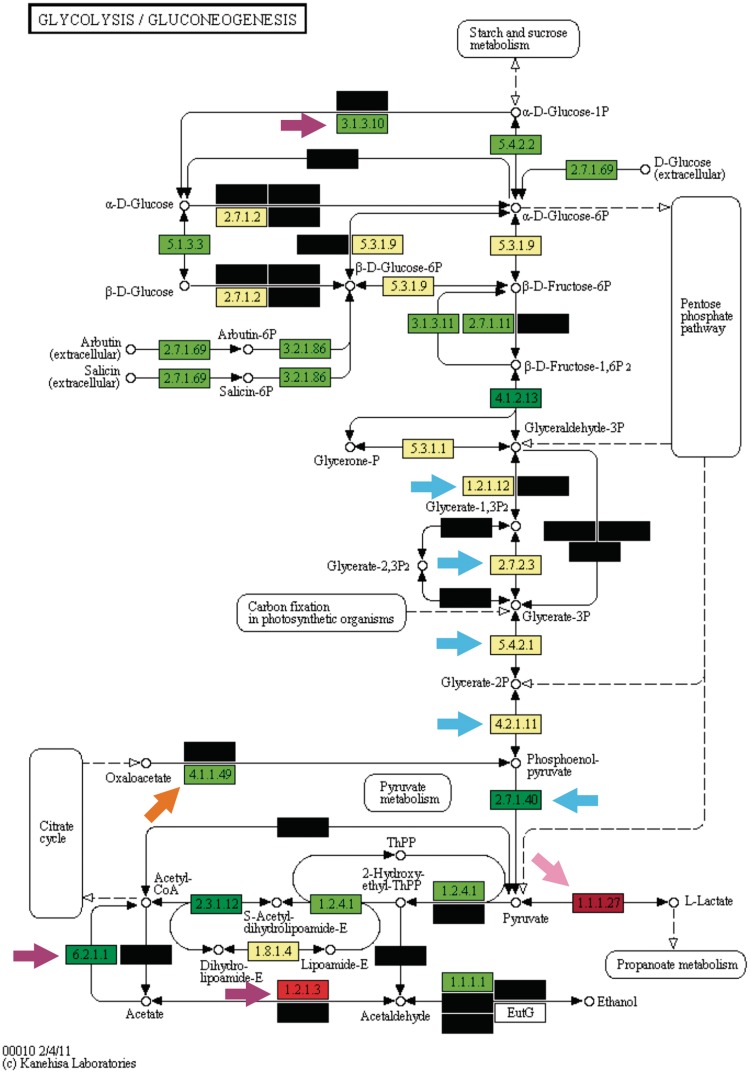
Figure 2.
KEGG Orthology enrichment analysis of the glycolytic pathway in the ENT group. Yellow boxes denote KOs where no difference was observed between test and background taxa. Black boxes denote KOs not observed in test or background taxa. Green and red boxes denote KOs significantly more and less represented in the test lineage when compared with the background one, respectively. Darker and lighter tones of both green and red boxes denote q-values between 0.05 and 0.00001 and smaller than 0.00001, respectively. Blue, pink, orange and purple arrows indicate trunk glycolysis enzymes, l-lactate dehydrogenase, PEP carboxykinase and taxon-specific enzymatic activities, respectively.
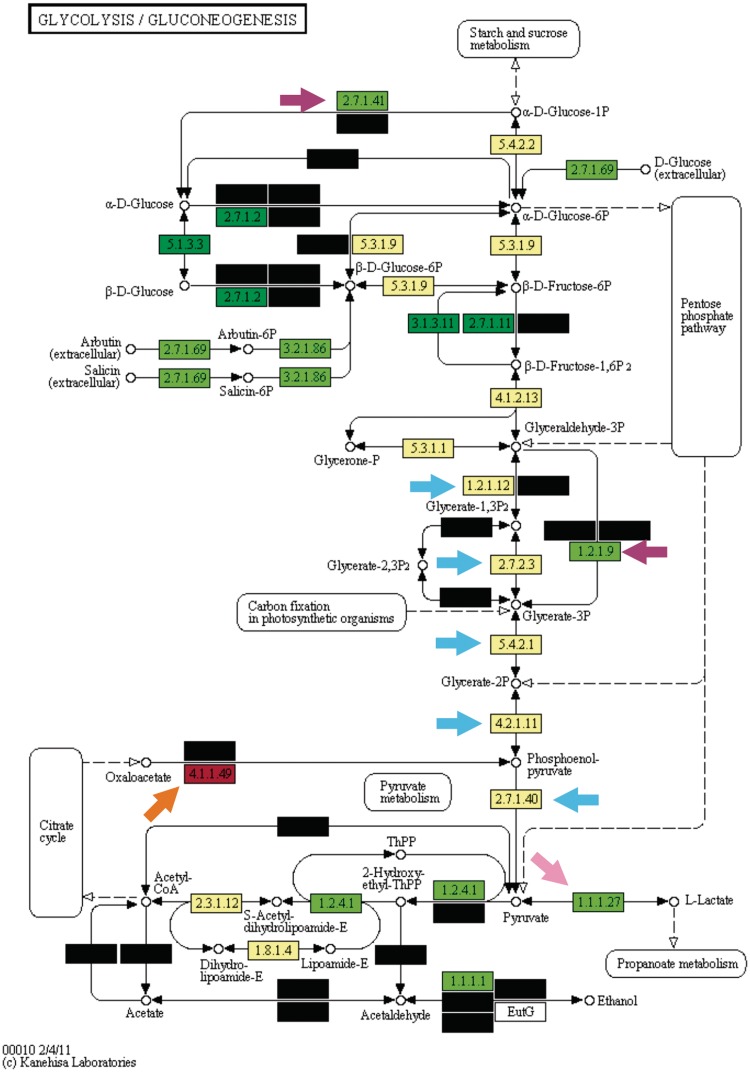
Figure 3.
KEGG Orthology enrichment analysis of the glycolytic pathway in the LAB group. The color schema and q-value cut-offs are the same as utilized in Figure 2.
Trunk glycolysis
Glycolysis is conceptually divided into a trunk portion and its lower and upper parts (12). Trunk glycolysis comprises the shared portion of the Embden–Meyerhof–Parnas, the Entner–Doudoroff and the pentose phosphate pathways, and converts glyceraldehyde 3-phosphate to pyruvate (Figures 2 and 3, blue arrows). This set of five enzymes provides key three-carbon intermediates to several anabolic pathways, including the biosynthesis of amino acids and fatty acids and is possibly the oldest portion of glycolysis, being shared by the three Domains of Life (12). The pattern we observed in our results supports this fact, since the trunk glycolysis enzymatic activities displayed practically no significant differences in either test taxa. Additionally, all trunk glycolytic enzymes were very abundant in the two test taxa as well as in the background taxon, with frequencies always >0.9. An interesting taxon-specific metabolic trait was detected in the trunk glycolysis: the bypass step catalyzed by the enzyme glyceraldehyde-3-phosphate dehydrogenase NADP+ dependent, which was strongly over-represented in the LAB group (Figure 3, EC:1.2.1.9, lower purple arrow). This bypass step generates a molecule of NADPH, an essential molecule for several anabolic pathways that is usually produced in pentose-phosphate pathway or in photosynthesis, but not commonly produced in glycolysis (10). This unusual finding led us to investigate the pentose-phosphate in the LAB group, where the key enzymes transketolase (EC:2.2.1.1) and transaldolase (EC:2.2.1.2) were under-represented in this taxon (Supplementary File 1, Supplementary Figure S1). The ENT group, on the other hand, possessed no such bias (Supplementary File 1, Supplementary Figure S2). Together, our findings suggest that the LAB group uses the bypass step observed in glycolysis to produce NADPH, a phenotype already suggested by in vivo studies (13).
Upper glycolysis
Very much unlike trunk glycolysis, the upper portion of glycolysis contained several enzymatic steps significantly over-represented; most of them common to both test taxa. Of special interest was the phosphotransferase (PTS) system, a group of several KOs strongly over-represented in both taxa (EC:2.7.1.69). The PTS system is a sugar transport mechanism composed of distinct groups of homologous genes that are widespread in several bacterial clades (14). Previous studies in fact suggest that this sugar transport system appears to be correlated with the niche occupied by the organisms where it is observed, with bacteria living in sugar-rich environments tending to possess significantly more distinct PTS genes (14). Together, the common over-represented enzymatic activities at upper glycolysis suggest that the two test taxa are able to perform several sugar transformation steps at the beginning of glycolysis, a finding consistent with their known metabolic niche (sugar-rich environments).
Lower glycolysis/Citrate cycle
Similarly to the pattern observed at the upper portion of glycolysis, the lower portion of this pathway also presented a considerable plasticity in the two test taxa, but with far more differences among them when compared with the upper portion. At the lower portion, we observed the only two enzymatic activities with opposite and inverted trends when comparing the test taxa, a finding highly correlated with the distinct fates of pyruvate in the ENT and LAB groups. The first of these trends was observed in the enzyme lactate dehydrogenase, which was under-represented in the ENT group and strongly over-represented in the LAB group (EC:1.1.1.27, pink arrows in Figures 2 and 3, respectively). The other divergent and inverted trend is the enzyme phosphoenolpyruvate (PEP) carboxykinase, which was strongly over-represented in the ENT group and under-represented in the LAB group (Figures 2 and 3, orange arrows, EC:4.1.1.49). The lactate dehydrogenase trend is a consequence of pyruvate commitment with lactate production in virtually all LAB members as well as in a few ENT members. The PEP carboxykinase enzyme is a known starting point to gluconeogenesis, given that it fuels this pathway with oxaloacetate to generate PEP.
One of the main sources of oxaloacetate in cellular metabolism is the citrate cycle, which suggested that the opposite pattern observed in PEP carboxykinase is due to possible differences in the citrate cycle in the two test taxa under analysis. When analyzing the citrate cycle pathway in ENT and LAB groups this was indeed the case (Supplementary File 1, Supplementary Figures S3 and S4, respectively). The ENT group possessed a complete version of this pathway, with most of the enzymatic steps being strongly over-represented. This pattern is expected due to the known capability of ENT members to fully oxidize pyruvate to CO2 through this pathway. The LAB group, on the other hand, possessed an incomplete version of this pathway, with most enzymatic activities strongly under-represented and a clear split of the citrate cycle into its oxidative and reductive branches (10). The partial citrate cycle found in the LAB group is related to the few members known to possess only an incomplete version of this pathway to fuel the biosynthesis of amino acids (8,9). Overall, the results depicted at the lower portion of glycolysis and in the citrate cycle are complementary and detected some of the key metabolic differences of the ENT and LAB groups regarding the distinct fates of pyruvate. At the lower glycolysis, it was also observed one ENT-specific enzymatic activity linked to a widely known phenotypical trait of this taxon. A key metabolical ability of Escherichia coli and other ENT members is their capability to grow utilizing citrate as the only carbon source (15). The enzymatic activity for this phenotype is due to the acetyl-CoA synthetase, which was found to be over-represented in this test taxon (Figure 2, lower purple arrow, EC:6.2.1.1).
CONCLUDING REMARKS
Complete genomes publicly available are now numbering in the thousands, and many more are expected to appear with the widespread use of next-generation DNA sequencing technologies (16). Additionally, a great effort is concomitantly underway to produce deep annotation information of distinct genomes to the same basic functional elements, such as the distinct coding regions, groups of homologous genes, operons and promoters, each of them associated to several dictionaries of biological roles (17). Consequently, the vast number of complete genomes available can be divided in groups based on biological criteria, such as taxonomic or environmental classification, and surveyed for the detection of biased distribution of distinct genomic components across them. Recently, several studies have proposed distinct methodologies to detect significantly biased biological roles as a function of the biological niche where genomic sequences have been sampled, a tendency induced by the maturation of the use of next-generation sequencing technologies to survey the genomic composition of distinct environments on earth (18,19). On the other hand, there have been virtually no previous attempts to develop a software that allows users to systematically look for significant distribution bias of biological features across the distinct existing taxa. It is widely accepted nowadays that several prokaryotic taxa higher than species possess ecological coherence. In other words, members of a taxon share general life strategies or traits that distinguish them from members of other taxa. Some obvious examples are the phylum Cyanobacteria (all known members are photoautotrophic) or the phylum Chlamydiae (all known members are obligate intracellular symbionts). Therefore, the study of the distribution of groups of homologous genes in higher order taxa can detect important properties to help understanding ecological traits of microbial communities (20). We believe KOMODO inaugurates the use of enrichment analysis as a statistical tool to systematically survey the distribution of shared genomic elements in distinct taxa in a quantitative way, making it possible to investigate how speciation shaped the genomic content of distinct taxa. The natural extension of this work will be the development of more general software to allow users to survey other genomic components significantly biased in a given group of genomes when compared with another by integrating the information available in the growing number of new genomic databases that are populating the field of computational biology (21).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–4, Supplementary File 1 and Supplementary References [22–26].
FUNDING
Funding for open access charge: Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq); KEGG database, Empresa Brasileira de Pesquisa Agropecuária (Embrapa).
Conflict of interest statement. None declared.
REFERENCES
- 1.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cai Z, Mao X, Li S, Wei L. Genome comparison using Gene Ontology (GO) with statistical testing. BMC Bioinformatics. 2006;7:374. doi: 10.1186/1471-2105-7-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weng MP, Liao BY. MamPhEA: a web tool for mammalian phenotype enrichment analysis. Bioinformatics. 2010;26:2212–2213. doi: 10.1093/bioinformatics/btq359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McLeay RC, Bailey TL. Motif enrichment analysis: a unified framework and an evaluation on ChIP data. BMC bioinformatics. 2010;11:165. doi: 10.1186/1471-2105-11-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T, et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36:D480–D484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Edwards PR, Ewing WH. Edwards and Ewing's Identification of Enterobacteriaceae. 4th edn. New York: Elsevier; 1986. [Google Scholar]
- 8.Ajdic D, McShan WM, McLaughlin RE, Savic G, Chang J, Carson MB, Primeaux C, Tian R, Kenton S, Jia H, et al. Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc. Natl Acad. Sci. USA. 2002;99:14434–14439. doi: 10.1073/pnas.172501299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wegmann U, O'Connell-Motherway M, Zomer A, Buist G, Shearman C, Canchaya C, Ventura M, Goesmann A, Gasson MJ, Kuipers OP, et al. Complete genome sequence of the prototype lactic acid bacterium Lactococcus lactis subsp. cremoris MG1363. J. Bacteriol. 2007;189:3256–3270. doi: 10.1128/JB.01768-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim BH, Gadd GM. Bacterial physiology and metabolism. Cambridge, NY: Cambridge University Press; 2008. [Google Scholar]
- 11.Lehninger AL, Nelson DL, Cox MM. Lehninger Principles of Biochemistry. 5th edn. New York: W.H. Freeman; 2008. [Google Scholar]
- 12.Storey KB. Functional Metabolism: Regulation and Adaptation. Hoboken, NJ: Wiley; 2004. [Google Scholar]
- 13.Asanuma N, Hino T. Presence of NAD+-specific glyceraldehyde-3-phosphate dehydrogenase and CcpA-dependent transcription of its gene in the ruminal bacterium Streptococcus bovis. FEMS Microbiol. Lett. 2006;257:17–23. doi: 10.1111/j.1574-6968.2006.00111.x. [DOI] [PubMed] [Google Scholar]
- 14.Barabote RD, Saier MH., Jr Comparative genomic analyses of the bacterial phosphotransferase system. Microbiol. Mol. Biol. Rev. 2005;69:608–634. doi: 10.1128/MMBR.69.4.608-634.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oh MK, Rohlin L, Kao KC, Liao JC. Global expression profiling of acetate-grown Escherichia coli. J. Biol. Chem. 2002;277:13175–13183. doi: 10.1074/jbc.M110809200. [DOI] [PubMed] [Google Scholar]
- 16.Neafsey DE, Haas BJ. 'Next-generation' sequencing becomes ‘now-generation’. Genome Biol. 2011;12:303. doi: 10.1186/gb-2011-12-3-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Muers M. Functional genomics: the modENCODE guide to the genome. Nat. Rev. 2011;12:80. doi: 10.1038/nrg2942. [DOI] [PubMed] [Google Scholar]
- 18.Gianoulis TA, Raes J, Patel PV, Bjornson R, Korbel JO, Letunic I, Yamada T, Paccanaro A, Jensen LJ, Snyder M, et al. Quantifying environmental adaptation of metabolic pathways in metagenomics. Proc. Natl Acad. Sci. USA. 2009;106:1374–1379. doi: 10.1073/pnas.0808022106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu H, Moore E. Association analysis of the general environmental conditions and prokaryotes' gene distributions in various functional groups. Genomics. 2010;96:27–38. doi: 10.1016/j.ygeno.2010.03.007. [DOI] [PubMed] [Google Scholar]
- 20.Philippot L, Andersson SG, Battin TJ, Prosser JI, Schimel JP, Whitman WB, Hallin S. The ecological coherence of high bacterial taxonomic ranks. Nat. Rev. Microbiol. 2010;8:523–529. doi: 10.1038/nrmicro2367. [DOI] [PubMed] [Google Scholar]
- 21.Bauer-Mehren A, Furlong LI, Sanz F. Pathway databases and tools for their exploitation: benefits, current limitations and challenges. Mol. Syst. Biol. 2009;5:290. doi: 10.1038/msb.2009.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ganzle MG, Vermeulen N, Vogel RF. Carbohydrate, peptide and lipid metabolism of lactic acid bacteria in sourdough. Food Microbiol. 2007;24:128–138. doi: 10.1016/j.fm.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 23.Dandekar T, Schuster S, Snel B, Huynen M, Bork P. Pathway alignment: application to the comparative analysis of glycolytic enzymes. Biochem. J. 1999;343 (Pt 1):115–124. [PMC free article] [PubMed] [Google Scholar]
- 24.Verhees CH, Kengen SW, Tuininga JE, Schut GJ, Adams MW, De Vos WM, Van Der Oost J. The unique features of glycolytic pathways in Archaea. Biochem. J. 2003;375:231–246. doi: 10.1042/BJ20021472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huynen MA, Dandekar T, Bork P. Variation and evolution of the citric-acid cycle: a genomic perspective. Trends Microbiol. 1999;7:281–291. doi: 10.1016/s0966-842x(99)01539-5. [DOI] [PubMed] [Google Scholar]
- 26.Arai K, Kamata T, Uchikoba H, Fushinobu S, Matsuzawa H, Taguchi H. Some Lactobacillus L-lactate dehydrogenases exhibit comparable catalytic activities for pyruvate and oxaloacetate. J. Bacteriol. 2001;183:397–400. doi: 10.1128/JB.183.1.397-400.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]