Abstract
We present a web-based tool set Rtips for fast and accurate prediction of RNA 2D complex structures. Rtips comprises two computational tools based on integer programming, IPknot for predicting RNA secondary structures with pseudoknots and RactIP for predicting RNA–RNA interactions with kissing hairpins. Both servers can run much faster than existing services with the same purpose on large data sets as well as being at least comparable in prediction accuracy. The Rtips web server along with the stand-alone programs is freely accessible at http://rna.naist.jp/.
INTRODUCTION
RNAs are versatile molecules for biological processes, working as messengers, regulators or catalysts in living cells. In particular, considerable attention has been focused on functions of regulatory non-coding RNAs. It is widely believed that there is a strong correlation between the 3D structure of an RNA molecule and its function. Since experimental determination of RNA 3D structure is difficult and their structures are hierarchical, computational prediction of secondary structures from a given single sequence or multiple sequences provides a major key to elucidating the potential functions of RNAs. Furthermore, interaction with another RNA or protein is often necessary for functional RNAs to perform their programmed tasks, and prediction of interacting structures is also an important problem in bioinformatics.
Taking as input either a single RNA sequence or a pair of RNA sequences, major software seeks to find an optimal secondary structure under a certain scoring function, given that the predicted structure has no complex motifs such as pseudoknots in intramolecular base pairings and kissing hairpins in intermolecular bindings. More specifically, a pseudoknot is typically formed from the base pairings between the unpaired bases of a loop and those outside the loop, whereas a kissing hairpin is caused by loop–loop interaction between two hairpin-type RNAs. Example predictive web tools are mfold (1), RNAfold (2) and CentroidFold (3) for RNA secondary structure prediction, and PairFold (4), RNAhybrid (5) and IntaRNA (6) for RNA–RNA interaction prediction. One reason why the complex motifs are disregarded is that the capability of handling such structural motifs results in high computational cost. However, it is observed that not a few number of these motifs occur in living cells, and thus these motifs should be considered in prediction algorithms to achieve more accurate prediction and avoid missing potential RNA genes in genome-wide sequence analysis. To this end, researchers have developed several tools together with web servers that can explicitly deal with such complicated motifs at the cost of computational efficiency such as NUPACK (7) and pknotsRG (8) for predicting secondary structures with pseudoknots, and inteRNA (9) for predicting RNA–RNA interactions with kissing hairpins. To summarize, it is desirable to clear the trade-off between the efficiency of a prediction algorithm and the class of predictable structures in order to broaden its applications.
To address this challenging problem, we have recently proposed two novel prediction methods, IPknot (10) for RNA secondary structure prediction including pseudoknots and RactIP (11) for RNA–RNA, interaction prediction including kissing hairpins, both of which employ integer programming (IP). Experimental validations of IPknot and RactIP indicate that our prediction methods are sufficiently accurate and quite fast even on large data sets as compared with several state-of-the-art methods [see (10,11)]. For easy access and use of those tools, we develop Rtips, a web server for Rna sTructure prediction using IP Scheme that comprises IPknot and RactIP. The website is free and open to all users, and there is no login requirement.
METHOD OVERVIEW
The methodology common to IPknot and RactIP is to combine the following two procedures when an RNA sequence or a pair of RNA sequences is given:
approximate a posterior probability distribution over a space of complex structures by its factorization;
maximize expected accuracy of a predicted structure by solving the corresponding IP problem.
In approximation of the probability distribution, we aim to decompose it into the product of probabilities defined over smaller base-pairing components, which are computationally easier to handle. The approximate probability distribution, explicitly represented as base-pairing posterior probabilities in the model, is incorporated into the objective function of the IP problem to find a secondary structure with the maximum expected accuracy (MEA). Expected accuracy can be expressed as the expected number of true predictions measured in base pairs. The IP problem is solved by GNU Linear Programming Kit (GLPK) in the web servers, which is freely available software for solving optimization problems. The advantage of using IP formulation is not only its strong descriptive power but also its flexibility and extensibility. In the framework of computing MEA, it is no longer necessary to consider the base pairs that do not contribute to improve expected accuracy, and thus we can take no account of them in advance.
The combination of the above procedures produces drastic speed-up in running time as well as good prediction accuracy. Therefore, the use of this strategy is very powerful to perform prediction even for large RNA sequences with complex motifs. Further details of our methodology can be found in (10,11).
GENERAL REMARKS
The top page of the Rtips web server provides links to respective web-based prediction services together with those to their source codes for stand-alone use and template programs to access the web services.
Each server accepts input by either entering RNA sequences directly or uploading FASTA files. The web interface has several optional parameters that affect prediction results. If the user does not adjust the parameters, the default values will be submitted to the server. Note that the default parameters related to calculating MEA (weight for true base pairs) were determined to obtain good predictions on many data sets and adjustment is hardly needed. Base-pairing posterior probabilities used in both tools are computed by RNAfold with parameters estimated by a Boltzmann likelihood-based method (12), which is based on McCaskill's dynamic programming algorithm (13) and thus we call it the McCaskill model, or by part of CONTRAfold (14), which is a machine learning-based predictor. If an illegal input is submitted to the server, the user will be notified of the inconsistency promptly. Each web interface for input includes automatic loading of several sample data to grasp the behavior of the tool, and provides interpretation of the output in the help page. It should be noted that we limit the size of input data to avoid overloading the servers, and the details of the restriction can be found in the help page of each server. If the size of submitted data is over the designated limit, the user is recommended to run the stand-alone program instead. If the user would like to integrate the functions of our servers with other web services, the template programs will be helpful.
After the job is submitted to the server, a prediction result can be found if the input is compatible. The result can be returned very quickly if the length of the submitted sequence is <400 nt. The user first finds a predicted 2D structure in dot-bracket representation, which can also be downloaded in Vienna format (2). To make the result easier to see, the server provides another graphical representation generated by VARNA (15). These graphics are embedded in the result page as PNG format, and those of original size are also available as PDF files.
IPKNOT SERVER
Input
The input is either a single RNA sequence or a multiple alignment of RNA sequences. If the user would like to know a secondary structure of a single RNA sequence, the sequence can be entered in plain or FASTA format into the field. Instead, the user can submit sequence information by uploading the corresponding FASTA file. Note that the length of the sequence must be at most 1500 nt. IPknot can also accept a multiple alignment of RNA sequences in CLUSTAL W format or multiple FASTA format to predict their consensus secondary structure. In this case, the alignment length is limited to 1500 nt. When pressing the ‘Predict’ button, the user can get a prediction result in the new page.
There are several parameters that IPknot can adjust. Level is the number of decompositions of a secondary structure where each decomposed substructure must have no pseudoknots. In other words, level can be considered as the number of kinds of brackets for indicating base pairs in dot-bracket representation. For example, level 1 uses just one kind of bracket ‘( )’, level 2 uses two kinds of brackets ‘( )’ and ‘[ ]’, and in level 3, three kinds of brackets ‘( )’, ‘[ ]’ and “{ }” are used. Therefore, IPknot of level 1 is an ordinary secondary structure predictor that does not consider pseudoknots like mfold and RNAfold, and it is almost equivalent to CentroidFold. IPknot of level 2 aims to predict nested pseudoknots, whereas IPknot of level 3 seeks to predict pseudoknotted structures with nested pseudoknots. The server provides three kinds of scoring models that produce base-pairing posterior probabilities. The McCaskill and the CONTRAfold models take no account of pseudoknotted structures in each decomposed substructure of IPknot, whereas the NUPACK model considers a certain class of pseudoknots in each substructure. Accordingly, the NUPACK model can be more accurate than the other two models to predict pseudoknotted secondary structures. However, a sequence of length >80 nt is too long for the elaborate NUPACK model to predict fast, and the server rejects the input. Besides, the NUPACK model is not supported for alignment input due to the computational cost. The user can choose whether the base-pairing probabilities of the McCaskill and CONTRAfold models defined over pseudoknot-free structures are refined or not. In the refinement procedure, the base-pairing probabilities are recalculated using the prediction result of the first run of IPknot. It should be noted that the choice of the NUPACK model disallows the refinement due to the computational cost of its iterative use. The weights of arbitrary positive numbers for respective levels can be specified in the web interface. Specifically, they represent the rate of true base pairs in the predicted secondary structure, which determine prediction accuracy. In general, if the weight increases, the algorithm aims to predict more base pairs and sensitivity of a prediction will get better. On the other hand, if the weight decreases, the algorithm tries to predict less base pairs and positive predictive value (PPV) will be enhanced. In this sense, the weights are balanced parameters between sensitivity and PPV.
Output
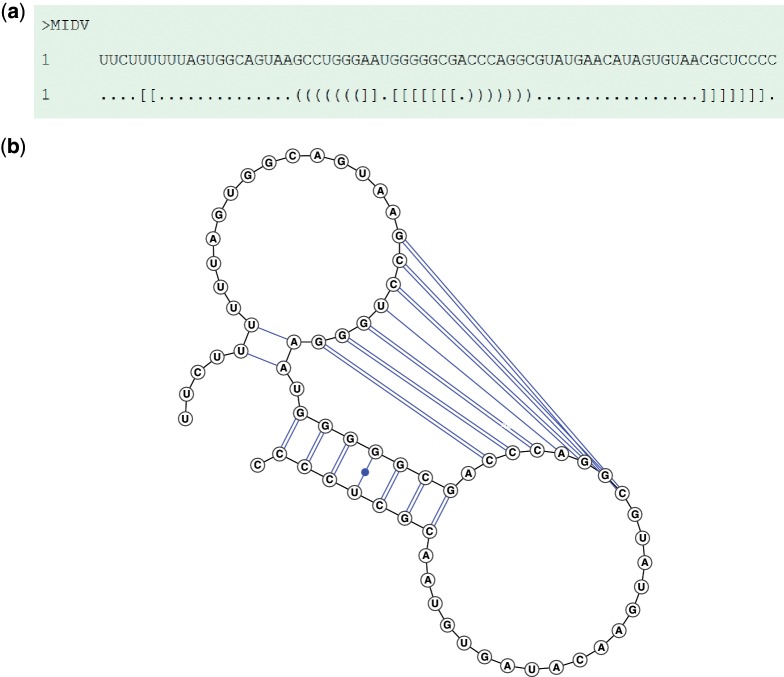
The user can find a predicted secondary structure with MEA. Figure 1a shows an example of a predicted MEA structure in dot-bracket representation where matching brackets indicate a base pair. Note that different forms of brackets, say ‘( )’ and ‘[ ]’ cross each other, meaning that the predicted structure includes pseudoknots. In addition to a downloadable Vienna file, the server can generate a BPSEQ formatted file for base-pairing information. A 2D diagram of the predicted structure along with its arc representation is displayed by running the VARNA program in the background [see Figure 1b]. Note that in the 2D diagram, an A–U pair is indicated by a single line with a bullet, a G–C pair is shown by a double line and a G–U pair is represented by a single line. In the result page for consensus structure prediction, the user can get the input alignment followed by the MEA common secondary structure in dot-bracket representation (Figure 2). Furthermore, a file that contains the predicted consensus structure as well as all input sequences in FASTA format is also downloadable. Interpretation of the other figures of a predicted structure is the same as that of a single sequence.
Figure 1.
Screenshot of the result page produced by the IPknot server when a sample sequence is submitted. The ‘MIDV’ sequence shown above is the 6K/TF ribosomal frameshift site of Middelburg virus, which was taken from PseudoBase (16). (a) Dot-bracket representation. (b) 2D diagram.
Figure 2.
Part of the result page when a multiple sequence alignment is submitted to the IPknot server. The sample alignment of tRNA-like structures was taken from Rfam 10.1 (17).
Validation
We validated prediction performance of IPknot on various data sets. One example of predicting a structure of a single sequence is a test on 388 non-redundant sequences of length at most 500 nt with at least one pseudoknot, showing 0.567, 0.578 and 0.570 in sensitivity, PPV and Matthews correlation coefficient (MCC), respectively, on average. Although these values may seem small, this is the best prediction performance as compared with other seven competitive methods [see (10)]. Another test on 67 alignments containing five sequences for consensus pseudoknotted structure prediction indicates 0.706, 0.717 and 0.707 in average sensitivity, PPV and MCC, respectively. An example of computation time is 3.95 s on a single sequence of length 989 nt, which was measured on the Linux machine identical to the web server (see the Implementation section for specifications). From the detailed validations in (10), IPknot is quite fast and sufficiently accurate as compared with several state-of-the-art methods.
RACTIP SERVER
Input
The input is a pair of RNA sequences in plain or FASTA format. Notice that each sequence must be put in 5′−3′ direction. Instead, the user can submit sequence information by uploading two separate FASTA files. Note that the sum of the lengths of two sequences must be at most 1000 nt, otherwise the server rejects the input. The user can get a prediction result in the new page by pressing the ‘Predict’ button.
The RactIP server offers two options. It provides the two aforementioned scoring models named CONTRAfold and McCaskill that produce internal base-pairing probabilities. In contrast, hybridization probabilities related to external base pairs are calculated by a variant of RNAduplex in the Vienna RNA package with parameters estimated by the Boltzmann likelihood-based method (12). Although the distinct models are used to derive internal base-pairing probabilities and hybridization probabilities, the approximation of the probability distribution enables us to select models separately that yield good predictions. Prediction accuracy depends on the specified weights as in the case of IPknot.
Output
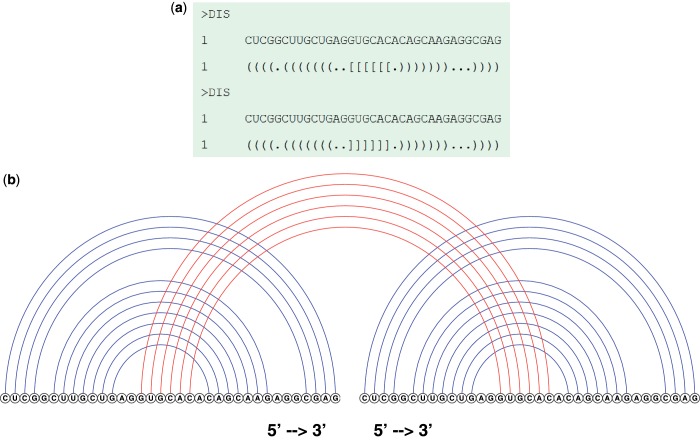
The output is a predicted joint secondary structure with MEA. The MEA structure is first shown in dot-bracket representation, where round brackets ‘( )’ indicate an internal base pair and square brackets ‘[ ]’ denote an external base pair (binding site) [see Figure 3a]. We should draw attention to the fact that there are no internal pseudoknots and external crossing interactions in joint structures predicted by RactIP, which is due to the assumption in the model. The free energy of the predicted joint secondary structure is given by employing RNAeval in the Vienna RNA package. A drawing of the predicted joint structure in arc representation is displayed, where blue arcs represent internal base pairs, red arcs stand for external interactions, and ‘5′ → 3′’ at the bottom shows the orientation of each RNA sequence [see Figure 3b].
Figure 3.
A sample output produced by the RactIP server. The above ‘DIS–DIS’ pair is caused by interaction between the partially self-complementary loops of the dimerization initiation sites of the HIV-1 genomic RNA (18). (a) Dot-bracket representation. (b) Arc representation.
Validation
We tested on 23 known RNA–RNA interaction pairs with total length of two sequences at most 300 nt. Five pairs out of 23 that are known to include kissing hairpins were used to evaluate the accuracy of predicted joint structures, indicating 0.963, 0.873 and 0.913 in sensitivity, PPV and MCC, respectively, on average. Looking at binding sites to assess the prediction accuracy on 23 RNA pairs, RactIP yields 0.833, 0.885 and 0.852 in average sensitivity, PPV and MCC, respectively. An example of running time measured on the machine described above is 0.855 s on an RNA–RNA pair of total lengths 306 nt. Detailed validations shown in (11) demonstrate that RactIP is extremely fast and sufficiently accurate as compared with several competitive prediction methods.
IMPLEMENTATION
The web server was implemented on a Linux CentOS 5 machine with Core i7-950 3.06 GHz CPU and 6.00 GB RAM using Apache, XHTML, JavaScript and PHP. The source codes for stand-alone use are written in C++, and the template programs to access the servers and parse the output are written in Perl.
DISCUSSION
The presented web tool set Rtips can predict sets of canonical base pairs from a set of input RNA sequences quite fast and accurately even if a secondary structure to be predicted is complicated. The proposed methods in Rtips are heuristic in the sense that they superimpose prediction results of primitive base-paired substructures to compose more complex secondary structures.
Other heuristic web tools that adopt the superimposition include HotKnots (19,20) for predicting secondary structures with pseudoknots and PETcofold (21,22) for predicting RNA–RNA interactions of multiple RNA sequences. IPknot is at least comparable in accuracy to HotKnots 2.0 (20) and can run an order of magnitude faster on large RNAs as shown in tests on various data sets in (10). The literature (21) reports that accuracy of RactIP is lower than that of PETcofold on condition that a set of homologous sequences is available, but running time of RactIP is much shorter. Equally importantly, RactIP needs no multiple alignment of RNA sequences that are expected to be homologous.
Our methodology will be powerful and useful enough to be applied to other important problems in RNA bioinformatics, including RNA structural alignment, prediction of non-canonical base pairs and genome-scale analysis associated with structure prediction. We have just got off to a good start to address these tasks and provide a potential extension of the server.
FUNDING
Grant-in-Aid for Young Scientists (B) from Japan Society for the Promotion of Science [#22700313 to Y.K., #22700305 to K.S.]. Funding for open access charge: Grant-in-Aid for Challenging Exploratory Research from Japan Society for the Promotion of Science [#23650153].
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors would like to thank all people who are involved in discussion about improvement on the server and testing the robustness.
REFERENCES
- 1.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gruber AR, Lorenz R, Bernhart SH, Neuböck R, Hofacker IL. The Vienna RNA Websuite. Nucleic Acids Res. 2008;36:W70–W74. doi: 10.1093/nar/gkn188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sato K, Hamada M, Asai K, Mituyama T. CentroidFold: a web server for RNA secondary structure prediction. Nucleic Acids Res. 2009;37:W277–W280. doi: 10.1093/nar/gkp367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andronescu M, Aguirre-Hernández R, Condon A, Hoos HH. RNAsoft: a suite of RNA secondary structure prediction and design software tools. Nucleic Acids Res. 2003;31:3416–3422. doi: 10.1093/nar/gkg612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Krüger J, Rehmsmeier M. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res. 2006;34:W451–W454. doi: 10.1093/nar/gkl243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith C, Heyne S, Richter AS, Will S, Backofen R. Freiburg RNA tools: a web server integrating IntaRNA, ExpaRNA and LocARNA. Nucleic Acids Res. 2010;38:W373–W377. doi: 10.1093/nar/gkq316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zadeh JN, Steenberg CD, Bois JS, Wolfe BR, Pierce MB, Khan AR, Dirks RM, Pierce NA. Software news and updates NUPACK: analysis and design of nucleic acid systems. J. Comput. Chem. 2011;32:170–173. doi: 10.1002/jcc.21596. [DOI] [PubMed] [Google Scholar]
- 8.Reeder J, Steffen P, Giegerich R. pknotsRG: RNA pseudoknot folding including near-optimal structures and sliding windows. Nucleic Acids Res. 2007;35:W320–W324. doi: 10.1093/nar/gkm258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aksay C, Salari R, Karakoc E, Alkan C, Sahinalp SC. taveRNA: a web suite for RNA algorithms and applications. Nucleic Acids Res. 2007;35:W325–W329. doi: 10.1093/nar/gkm303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sato K, Kato Y, Hamada M, Akutsu T, Asai K. IPknot: fast and accurate prediction of RNA secondary structures with pseudoknots using integer programming. Bioinformatics. 2011;27:i85–i93. doi: 10.1093/bioinformatics/btr215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kato Y, Sato K, Hamada M, Watanabe Y, Asai K, Akutsu T. RactIP: fast and accurate prediction of RNA–RNA interaction using integer programming. Bioinformatics. 2010;26:i460–i466. doi: 10.1093/bioinformatics/btq372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Andronescu M, Condon A, Hoos HH, Mathews DH, Murphy KP. Computational approaches for RNA energy parameter estimation. RNA. 2010;16:2304–2318. doi: 10.1261/rna.1950510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McCaskill JS. The equilibrium partition function and base pair probabilities for RNA secondary structure. Biopolymers. 1990;29:1105–1119. doi: 10.1002/bip.360290621. [DOI] [PubMed] [Google Scholar]
- 14.Do CB, Woods DA, Batzoglou S. CONTRAfold: RNA secondary structure prediction without physics-based models. Bioinformatics. 2006;22:e90–e98. doi: 10.1093/bioinformatics/btl246. [DOI] [PubMed] [Google Scholar]
- 15.Darty K, Denise A, Ponty Y. VARNA: interactive drawing and editing of the RNA secondary structure. Bioinformatics. 2009;25:1974–1975. doi: 10.1093/bioinformatics/btp250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Batenburg FHD, Gultyaev AP, Pleij CWA, Ng J, Oliehoek J. PseudoBase: a database with RNA pseudoknots. Nucleic Acids Res. 2000;28:201–204. doi: 10.1093/nar/28.1.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gardner PP, Daub J, Tate J, Moore BL, Osuch IH, Griffiths-Jones S, Finn RD, Nawrocki EP, Kolbe DL, Eddy SR. Rfam: Wikipedia, clans and the “decimal” release. Nucleic Acids Res. 2011;39:D141–D145. doi: 10.1093/nar/gkq1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paillart JC, Skripkin E, Ehresmann B, Ehresmann C, Marquet R. A loop–loop “kissing” complex is the essential part of the dimer linkage of genomic HIV-1 RNA. Proc. Natl. Acad. Sci. USA. 1996;93:5572–5577. doi: 10.1073/pnas.93.11.5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ren J, Rastegari B, Condon A, Hoos HH. HotKnots: heuristic prediction of RNA secondary structures including pseudoknots. RNA. 2005;11:1494–1504. doi: 10.1261/rna.7284905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Andronescu M, Pop C, Condon A. Improved free energy parameters for RNA pseudoknotted secondary structure prediction. RNA. 2010;16:26–42. doi: 10.1261/rna.1689910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seemann SE, Richter AS, Gesell T, Backofen R, Gorodkin J. PETcofold: predicting conserved interactions and structures of two multiple alignments of RNA sequences. Bioinformatics. 2011;27:211–219. doi: 10.1093/bioinformatics/btq634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seemann SE, Menzel P, Backofen R, Gorodkin J. The PETfold and PETcofold web servers for intra- and intermolecular structures of multiple RNA sequences. Nucleic Acids Res. 2011;39:W107–W111. doi: 10.1093/nar/gkr248. [DOI] [PMC free article] [PubMed] [Google Scholar]