Abstract
In this article, we introduce BioMe (biologically relevant metals), a web-based platform for calculation of various statistical properties of metal-binding sites. Users can obtain the following statistical properties: presence of selected ligands in metal coordination sphere, distribution of coordination numbers, percentage of metal ions coordinated by the combination of selected ligands, distribution of monodentate and bidentate metal-carboxyl, bindings for ASP and GLU, percentage of particular binuclear metal centers, distribution of coordination geometry, descriptive statistics for a metal ion–donor distance and percentage of the selected metal ions coordinated by each of the selected ligands. Statistics is presented in numerical and graphical forms. The underlying database contains information about all contacts within the range of 3 Å from a metal ion found in the asymmetric crystal unit. The stored information for each metal ion includes Protein Data Bank code, structure determination method, types of metal-binding chains [protein, ribonucleic acid (RNA), deoxyribonucleic acid (DNA), water and other] and names of the bounded ligands (amino acid residue, RNA nucleotide, DNA nucleotide, water and other) and the coordination number, the coordination geometry and, if applicable, another metal(s). BioMe is on a regular weekly update schedule. It is accessible at http://metals.zesoi.fer.hr.
INTRODUCTION
Metal cations are constituents of approximately 40% of all proteins (1), they take part in enzymatic reactions and they are the essential partners in assembly of the functional ribonucleic acid (RNA) structures (2,3). For example, presence of magnesium di-cations is essential for the formation and stabilization of the transfer RNA tertiary structure (4,5). A region around a metal often defines the so-called ‘active place’ where a particular chemical reaction can take place. Each metal ion possesses unique combination of charge and rigidity of its coordination sphere. Removal or replacement of one metal by another is accompanied by loss, reduction or even alteration of the enzyme catalytic potency [see for example (6)]. The knowledge of the metal ions environment, especially the electron donor types and number, is important to clarify how specific a metal-binding site is and how we can tune the desired chemical reaction.
To the best of our knowledge, this is the only website that focuses on the binding sites statistical properties. Currently, there are several databases available for researchers to view the information on metals and metal-binding sites in proteins [MetLigDB (7), MESPEUS (8), MIPS (9), MDB (10) and COMe (11)] and RNAs [MeRNA (12)]. However, they are typically limited to simple retrieval of Protein Data Bank (PDB) structures for specified metal and donor atoms.
The main contribution of BioMe is that, unlike the databases discussed earlier, it generates statistical reports for predefined or user-defined PDB subsets. This approach allows the users to find characteristics of particular metal-biding sites in a simple and straightforward manner. In addition, to the best of our knowledge, BioMe is the only website that distinguishes between various chain types [i.e. protein, RNA and deoxyribonucleic acid (DNA)], thus enabling the user to easily focus on the chain of their interest. Our website additionally provides information about the binuclear metal centers, the occurrence of certain combination of molecules (13) in metal ion coordination sphere and about the metal coordination geometry (14), for coordination numbers ranging from 3 to 14. Finally, the MySQL database dump of BioMe underlying database is publicly available. Among the existing databases, as far as we know, only MDB (the time of the last update was in 2003) has the dump available, whereas querying MESPEUS SQL database is available only on request (by sending an email to author).
The main motivations for setting up BioMe are exponentially growing number of three-dimensional (3D) structures deposited in the PDB (15), the green chemistry requirements for increasing efficiency of existing metaloenzymes and design of the new ones. This is a continuation of our earlier work on structure of the metal-binding sites in proteins (13). A user-friendly interface enables scientists to perform their own statistics and to easily extrapolate the relevant data. In addition, the website enables the retrieval of information concerning natural, as well as toxic metal cations, precisely the all metal ions found in PDB to be tightly bound either to a protein or/and to a nucleic acid are considered. In comparison with the other mentioned databases, BioMe enables queries for a multiple selection of metals and ligands. Furthermore, for the purpose of getting more general picture of the metal ions distribution in different types of proteins, the pre-calculated nonredundant PDB set is also available.
METHODS
BioMe underlying database is built from the 3D structures (PDB files) in which the metal ions are coordinated with at least either two donor atoms from a protein chain or alternatively one donor atom from a nucleic acid chain. The purpose of these constraints is try to eliminate metals added for the crystallization purposes. For each PDB entry, we have extracted the PDB code, title, information about the method used to derive the 3D structure, resolution and the release date. Distances between the metal ions and the electron donor atoms (O, N, S, Cl and F), as well as those between the metals ions themselves, are calculated from the atomic coordinates. In the cases where either the metal ion and/or some of its electron donors have multiple positions within the crystal structure, the position with higher occupancy was selected. In this study, we distinguish protein, RNA, DNA, ‘other’ chains and water. Protein chains and nucleic acid chains (RNA and DNA) are defined as chains with at least 50 amino acid residues and five nucleotides, respectively.
We use the distance limit of 3 Å for defining the coordination bond as in (13). This high threshold was used to account for the natural flexibility of a metal coordination sphere and for the possible coordination errors in structure determination. A metal coordination number was calculated by summing the number of all electronegative atoms within the 3 Å range (O, N, S, Cl and F). The calculation of the coordination geometry (14) is based on the geometrical pattern of the coordinating atoms L1, L2, … , Ln, which is described by the list W of all bond angles around the metal ion M sorted in ascending order:
In the same way, a set of angle lists of ideal coordination geometries was developed. The root mean square deviation (RMSD) is calculated between the angle list of the particular metal ion and each of the ideal geometry lists of the same length. The list that shows smallest RMSD is used as the best fitting coordination geometry (16). Currently, the web server calculates 22 different geometries for coordination numbers ranging from 4 to 14. Ideal coordination geometries used in this work are presented in Table 1.
Table 1.
Coordination geometries used in BioMe web server
| Coordination number | Geometry |
|---|---|
| 4 | Tetrahedron |
| 4 | Square planar |
| 5 | Trigonal bipyramid |
| 5 | Square pyramid (tetragonal bipyramid) |
| 6 | Octahedron |
| 6 | Trigonal prism |
| 7 | Octahedron, face capped |
| 7 | Trigonal prism, square face monocapped |
| 7 | Pentagonal bipyramid |
| 8 | Dodecahedron (bisdisphenoid) |
| 8 | Cube |
| 8 | Hexagonal bipyramid |
| 8 | Trigonal prism, square face bicapped |
| 8 | Square antiprism |
| 8 | Trigonal prism, triangular face bicapped |
| 9 | Square antiprism, monocapped |
| 9 | Trigonal prism, square face tricapped |
| 10 | Square antiprism, bicapped |
| 12 | Cuboctahedron |
| 12 | Anticuboctahedron |
| 12 | Icosahedron |
| 14 | Hexagonal antiprism, bicapped |
Although in our previous work (13), we used only a set of most representative metals, in this study, we have taken into account all the metals found in the PDB structures.
After parsing the whole PDB, we found 20 307 files with structures that satisfied our conditions. Among those, there are 43 326 protein chains, 650 DNA chains and 734 RNA chains. However, there are a number of redundant structures. Hence, for the purpose of getting an unbiased picture of the metal ions distribution in proteins, the list of metals in the nonredundant, representative selection of structures is also available. The representative set of the protein chains and the corresponding PDB files was extracted from the PDB (April 2012) according to a pre-calculated nonredundant set of ‘cluster-70 chains’ downloaded from the PDB site. From the each cluster, we chose the best-ranked chain and checked whether it contains a metal cation that satisfies defined conditions. If such a metal was not found in the best ranked chain, we chose the second one on the list and repeated the procedure recursively until we found a structure with a bound metal ion or reached the end of the list.
The database is implemented with MySQL on a Gentoo Linux operating system (Intel Core2 Quad CPU Q6600 at 2.40 GHz, 4 GB).
USER INTERFACE
The main purpose of the user interface is to create a query and to present the results (statistics) in an appropriate form. The starting page offers a large number of options, so users can easily tailor their queries. They can refine their search by type of the metal ions, the structure determination method (X-ray crystallography, NMR or both) and resolution, by the type of chain and combination of ligands, by the coordination number, maximum RMSD for a metal coordination geometry (the default value is 15°) and by the threshold distance between the selected metal ions. A help page exists for each parameter.
The search can be performed on the prepared list of all PDBs that fulfilled the requirements (metal ions with at least two donor atoms from a protein chain or at least one from a nucleic acid chain) and/or on the representative cluster-70 list. Moreover, users can narrow their search by specifying a list of structures.
Currently, users can select up to 25 most representative kinds of ions (Mg, Zn, Ca, Fe, Na, Mn, K, Sr, Cu, Cd, Ni, Hg, Co, W, Os, Mo, Ba, Al, Tl, Au, Pt, Pb, V, Yb and Sm). However, using the publicly available database dump, users can perform search for any metal available in the PDB.
BioMe distinguishes between five different types of ligands: amino acid residues, RNA and DNA nucleotides, water and ‘others’. In the output statistics, ligands belonging to the first four types are presented with their names, whereas those from the last group are presented just as ‘others’. We found that using the original names from PDB files for the last group of ligands introduces a poorly legible graphical representation of the results, especially when the search is performed for a large number of structures. Therefore, the results for them are available on a separate page that can be accessed by ‘Other list’ button.
Among the available results, we distinguish between two types of statistics: statistics associated with the selected metals and statistics associated with the selected amino acids and nucleotides.
For each metal, seven statistics are available (i) relative presence of selected ligands in the metal coordination sphere, (ii) relative distribution of the coordination numbers, (iii) percentage of metal ions with the selected coordination number coordinated by combination of selected ligands, (iv) distribution of monodentate and bidentate metal-carboxyl bindings for Asp and Glu, (v) number of particular binuclear metal centers, (vi) distribution of coordination geometries and (vii) average distances and standard deviations for selected metal and donor atoms.
The last statistics, Statistics 8, is performed if more than one metal is selected. For each metal ion, it gives information on how often it is coordinated with each of the selected ligands.
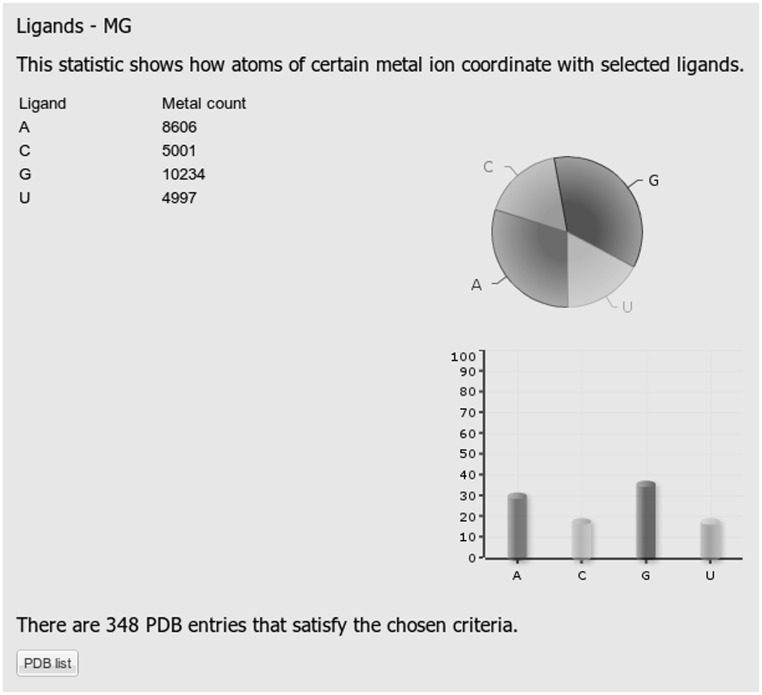
With the exception of Statistics 3 and Statistics 7, all the remaining statistics are presented numerically and graphically. Graphs include pie and column charts. As an illustration, Figure 1 shows a result for Statistics 1. In this example, the query includes Mg ions in any coordination number bound to any type of RNA nucleotide (in the RNA chain). As can be seen from Figure 1, Mg is mostly bound to G, followed by A and C and U base. Concurrent binding to G and A corresponds to binding to the shared G A pairs, as described in Stefan et al. (12) and references therein.
Figure 1.
A screenshot displaying results of statistics number 1 showing the number of each type of RNA nucleotide that binds to Mg. The statistics represents only nucleotides in RNA chains.
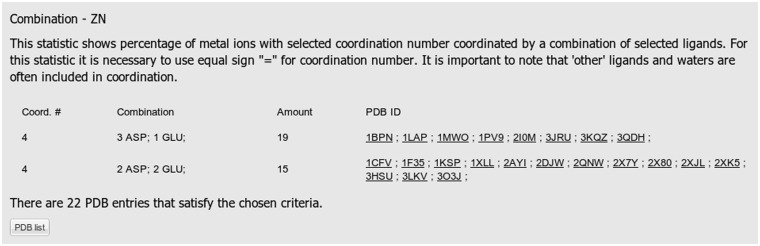
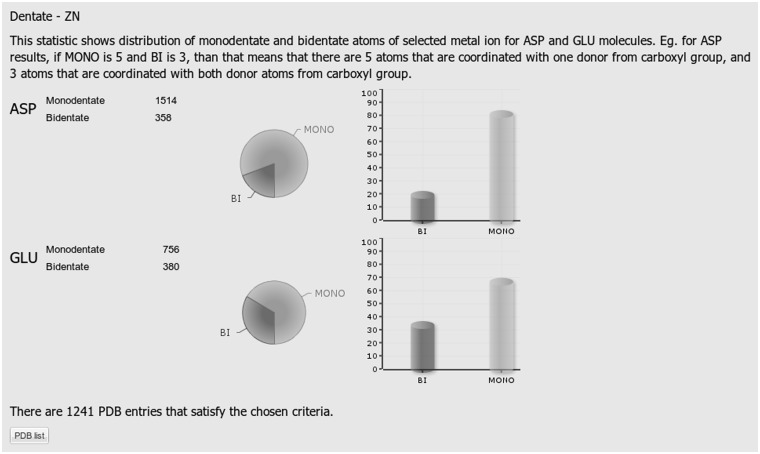
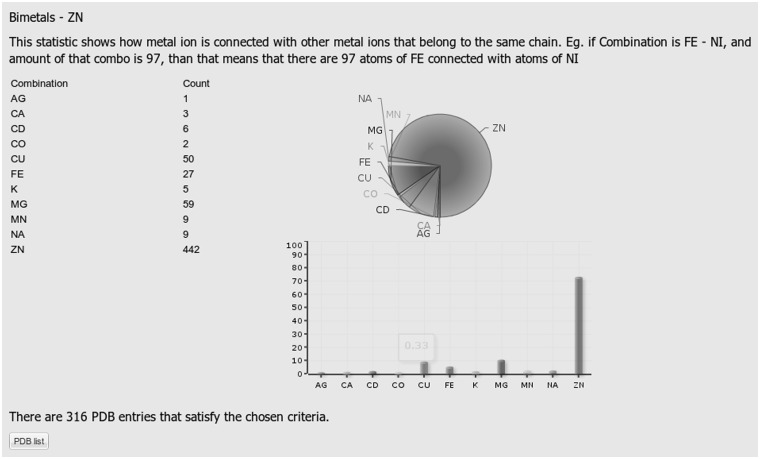
Examples of the outputs for Statistics 3, 4 and 5 are presented in Figures 2, 3 and 4, respectively. The query specified Zn as a metal of interest, Asp and Glu amino acid residues as ligand types, protein as a chain type, 4 as a desired coordination number and the entire database as a set of structures to be searched.
Figure 2.
A screenshot displaying results of statistics number 3 showing the number of the Zn ions with the coordination number 4 coordinated by the given combination of ligands. In the query, the following selections were made: entire PDB, Zn, Asp and Glu amino acid residues, protein chain type and coordination number 4.
Figure 3.
A screen shot of statistic number 4 (distribution of monodentate and bidentate metal-carboxyl bindings for Asp and Glu) obtained from a query that included all available PDB structures, Zn atom, Asp and Glu amino acid residues, protein chain type and coordination number 4.
Figure 4.
A screenshot displaying results of statistics number 5 which revealing the number of binuclear metal centers containing the Zn ion together with the percentage of metals that together with the zinc dication form a binuclear metal center. In the query, the following selections were made: entire PDB, Zn, Asp and Glu amino acid residues, protein chain type and coordination number 4.
The Statistics 4 classifies the carboxyl binding as monodentate, in the case where only one of its O atoms participates in the coordination of a particular metal ion and bidentate in the case where both oxygen atoms are involved in the metal coordination.
For the purpose of further processing, the results are available in the csv file format. Furthermore, for each of the statistics, there is a list of PDB files that satisfy the query with links to their entries in PDB.
The dump of the underlying database is publicly available in a separate window that can be accessed from the starting page, and it gives users the opportunity to perform their own analysis. The database is on a regular weekly update schedule.
The user interface was built as a web application using GWT development toolkit (http://code.google.com/webtoolkit/). Tomcat is used as the application server.
APPLICATION
The statistics obtained using the presented databases may help in the identification and modeling of the metal-binding sites in the protein structures derived by homology modeling and in design of proteins with a specific affinity for a certain metal (so-called metal biosorbents) with potential application in the environmental chemistry. Carefully designed and performed statistics could provide a clue about which metal ion and/or environment would be the best choice for a certain reaction. Thus, it should also help to improve catalytic performances of existing and aid the design of new ones.
CONCLUSIONS
The website allows scientists to perform a number of different searches and to obtain useful information about the selected metal ions in biological macromolecules and their ligands. Users can choose to retrieve information from all structures available in PDB, from a nonredundant set of protein chains or from their own list of structures. The search can be performed either for proteins or nucleic acids, RNAs and DNAs or both. For each selection, information about the coordination numbers, distances, percentage of monodentately and bidentately bound Asp and Glu carboxyl groups, percentage of metal ions with selected coordination number coordinated by combination of selected ligands, coordination geometry and population of particular binuclear metal centers users can retrieve. We believe that BioMe will prove a valuable tool for all research related to metal ions in proteins and nucleic acids.
FUNDING
Ministry of Education Science and Sports of the Republic of Croatia [098-1191344-2860 to S.T. and M.Š.] and [036-0362214-1987 to M.Š.]; Biomedical Research Council of A*STAR, Singapore to M.Š. Funding for open access charge: Biomedical Research Council of A*STAR, Singapore.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank Antonija Tomić for her help in testing the server and helpful comments on the interface functionality, Ivana Mihalek and Ana Bulović for proofreading the manuscript and Mak Krnić for the help in deploying the application.
REFERENCES
- 1.Thomson AJ, Gray HB. Bio-inorganic chemistry. Curr. Opin. Chem. Biol. 1998;2:155–158. doi: 10.1016/s1367-5931(98)80056-2. [DOI] [PubMed] [Google Scholar]
- 2.Woodson SA. Metal ions and RNA folding: a highly charged topic with a dynamic future. Curr. Opin. Chem. Biol. 2005;9:104–109. doi: 10.1016/j.cbpa.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 3.Zemora G, Waldsich C. RNA folding in living cells. RNA Biol. 2010;7:634–641. doi: 10.4161/rna.7.6.13554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leroy JL, Guéron M, Thomas G, Favre A. Role of divalent ions in folding of tRNA. Eur. J. Biochem./FEBS. 1977;74:567–574. doi: 10.1111/j.1432-1033.1977.tb11426.x. [DOI] [PubMed] [Google Scholar]
- 5.Misra VK, Shiman R, Draper DE. A thermodynamic framework for the magnesium-dependent folding of RNA. Biopolymers. 2003;69:118–136. doi: 10.1002/bip.10353. [DOI] [PubMed] [Google Scholar]
- 6.Leitgeb S, Nidetzky B. Enzyme catalytic promiscuity: the nonheme Fe2+ center of beta-diketone-cleaving dioxygenase Dke1 promotes hydrolysis of activated esters. Chembiochem: Eur. J. Chem. Biol. 2010;11:502–505. doi: 10.1002/cbic.200900688. [DOI] [PubMed] [Google Scholar]
- 7.Choi H, Kang H, Park H. MetLigDB: a web-based database for the identification of chemical groups to design metalloprotein inhibitors. J. Appl. Crystallogr. 2011;44:878–881. [Google Scholar]
- 8.Hsin K, Sheng Y, Harding MM, Taylor P, Walkinshaw MD. MESPEUS: a database of the geometry of metal sites in proteins. J. Appl. Crystallogr. 2008;41:963–968. [Google Scholar]
- 9.Hemavathi K, Kalaivani M, Udayakumar A, Sowmiya G, Jeyakanthan J, Sekar K. MIPS: metal interactions in protein structures. J. Appl. Crystallogr. 2009;43:196–199. [Google Scholar]
- 10.Castagnetto JM, Hennessy SW, Roberts VA, Getzoff ED, Tainer JA, Pique ME. MDB: the Metalloprotein Database and Browser at The Scripps Research Institute. Nucleic Acids Res. 2002;30:379–382. doi: 10.1093/nar/30.1.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Degtyarenko K, Contrino S. COMe: the ontology of bioinorganic proteins. BMC Struct. Biol. 2004;4:3. doi: 10.1186/1472-6807-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stefan LR, Zhang R, Levitan AG, Hendrix DK, Brenner SE, Holbrook SR. MeRNA: a database of metal ion binding sites in RNA structures. Nucleic Acids Res. 2006;34:D131–D134. doi: 10.1093/nar/gkj058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dokmanić I, Sikić M, Tomić S. Metals in proteins: correlation between the metal-ion type, coordination number and the amino-acid residues involved in the coordination. Acta Crystallogr. D Biol. Crystallogr. 2008;64:257–263. doi: 10.1107/S090744490706595X. [DOI] [PubMed] [Google Scholar]
- 14.Lima-de-Faria J, Hellner E, Liebau F, Makovicky E, Parth E. Nomenclature of inorganic structure types. Report of the International Union of Crystallography Commission on Crystallographic Nomenclature Subcommittee on the Nomenclature of Inorganic Structure Types. Acta Crystallogr. A Found. Crystallogr. 1990;46:1–11. [Google Scholar]
- 15.Berman HM. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seebeck B, Reulecke I, Kämper A, Rarey M. Modeling of metal interaction geometries for protein-ligand docking. Proteins. 2008;71:1237–1254. doi: 10.1002/prot.21818. [DOI] [PubMed] [Google Scholar]