Abstract
A module is a fundamental unit forming with highly connected proteins and performs a certain kind of biological functions. Modules and module–module interaction (MMI) network are essential for understanding cellular processes and functions. The MoNetFamily web server can identify the modules, homologous modules (called module family) and MMI networks across multiple species for the query protein(s). This server first finds module candidates of the query by using BLASTP to search the module template database (1785 experimental and 1252 structural templates). MoNetFamily then infers the homologous modules of the selected module candidate using protein–protein interaction (PPI) families. According to homologous modules and PPIs, we statistically calculated MMIs and MMI networks across multiple species. For each module candidate, MoNetFamily identifies its neighboring modules and their MMIs in module networks of Homo sapiens, Mus musculus and Danio rerio. Finally, MoNetFamily shows the conserved proteins, PPI profiles and functional annotations of the module family. Our results indicate that the server can be useful for MMI network (e.g. 1818 modules and 9678 MMIs in H. sapiens) visualizations and query annotations using module families and neighboring modules. We believe that the server is able to provide valuable insights to determine homologous modules and MMI networks across multiple species for studying module evolution and cellular processes. The MoNetFamily sever is available at http://monetfamily.life.nctu.edu.tw.
INTRODUCTION
A module is a fundamental unit forming with highly connected proteins and often possesses specific biological functions. The interactions between modules are considered as the backbone of the cellular networks to regulate most biological processes (BP) (1,2). To infer biological modules and module–module interaction (MMI) networks is an emergency task for understanding cellular processes. As an increasing number of complete genomes become available, identifying homologous modules and the MMIs provides an opportunity for inferring new modules and MMI networks across multiple species.
Many methods have been proposed to identify biological modules [e.g. functional (3,4) and evolutionary modules (5,6)] and few databases provided biological modules across multiple species (7–9). However, these studies are often lack of the relationships between modules. Recently, a systems biology view of modules and their MMIs has been proposed in a target organism, such as Homo sapiens (1) or Saccharomyces cerevisiae (2,10). These works showed that MMIs and a module network can be useful for analyzing biological functions and processes. However, these methods are often limited or time-consuming to identify homologous modules, MMIs and module networks across multiple species. The concept of the module family is analogous to the concepts of protein sequence family (11) and protein structure family (12) and protein–protein interactions (PPI) family (13). To infer homologous modules and MMI networks across multiple species from a large complete genomic database [e.g. 2274 species in Integr8 version 103 (14)] is an important issue for studying module evolution and cellular processes.
To address these issues, we constructed MoNetFamily, a server for identifying homologous modules (called module family) and MMIs in module networks across multiple species. According to our knowledge, MoNetFamily is the first public server that infers MMI networks in H. sapiens, Mus musculus and Danio rerio of the query proteins using homologous modules. For a set of query protein(s), this server provides homologous modules, graphic visualization of MMI networks and neighboring modules, PPI profiles and conserved Gene Ontology (GO) annotations (15).
METHOD AND IMPLEMENTATION
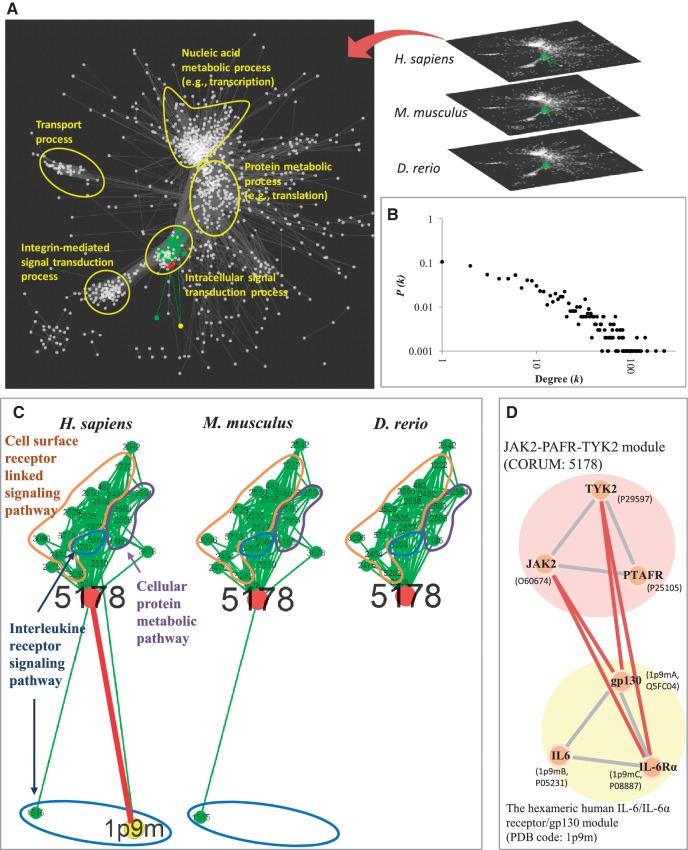
Figure 1 shows the details of the MoNetFamily server to search the template-based homologous modules and the MMI networks of a set of query protein sequence(s), gene name(s), or UniProtKB accession number(s) by the following steps (Figure 1A and Supplementary Figure S1): first, the server uses BLASTP to search module candidates from the module template database with the protein similarity (E-values ≤ 10−10) (16) (Figure 1B). This database consists of 1975 non-redundant modules with 4659 proteins. A total of 1785 protein complexes (three or more proteins) were selected from comprehensive resource of mammalian protein complexes database (CORUM; released on 02 September 2009) (9) and 1252 structure complexes were selected from Protein Data Bank (PDB; released on 25 December 2009) (17). For each module candidate, this server provides the neighboring modules and MMIs (Supplementary Figure S1C) in MMI networks of H. sapiens, M. musculus and D. rerio (Figures 1C and 2) through homologous modules. Next, the homologous modules of the template candidate are derived from the PPI families (13,18,19) (Figure 1B and D). According to homologous modules and PPIs, we statistically calculated MMIs (Figure 2D) and MMI networks across multiple species using the hypergeometric distribution. The MoNetFamily server consists of 1818, 1801 and 1586 modules in H. sapiens, M. musculus and D. rerio, respectively. In the MMI networks, 1440 (H. sapiens), 1396 (M. musculus) and 1257 (D. rerio) modules are interconnected. For a module family and its neighboring modules, we measured the consensus ratios and adjusted P-values of BP, cellular components (CC) and molecular functions (MF) based on GO annotations (Figure 1C). Finally, this server provides not only homologous modules but also graphic visualization of the neighboring modules and MMIs in the MMI networks across multiple vertebrates.
Figure 1.
Overview of the MoNetFamily server for MMI network and homologous module search using proteins Jak2, Ptafr and Tyk2 of M. musculus as the query. (A) Main procedure. (B) Input the query protein(s) and identify the candidates of the query using BLASTP to scan the module template database. (C) The neighboring modules and MMIs of the selected template (CORUM ID: 5178) in MMI networks of H. sapiens, M. musculus and D. rerio. The conserved GO annotations of the module family and neighboring modules of the query are indicated. (D) The profiles of proteins and PPIs in the selected homologous modules.
Figure 2.
The MMI networks of H. sapiens, M. musculus and D. rerio. (A) Five major cellular processes in MMI networks in H. sapiens. (B) The node degree distribution of MMI network in H. sapiens follows a power law. The MMI network is a scale-free network. (C) The neighboring module network between the JAK2–PAFR–TYK2 module (CORUM ID: 5178) and its 25 neighboring modules. These modules can be roughly divided into three groups, including cell surface receptor linked signaling pathway (orange), cellular protein metabolic pathway (purple) and interleukine receptor signaling pathway (blue). Neighboring modules are highly consensus on two GO terms that are signal transducer activity and cytosol. (D) The inter-module PPIs (red lines) and the topology between modules JAK2–PAFR–TYK2 and the hexameric human IL-6/IL-6α receptor/gp130 (PDB code: 1p9m).
Homologous module
Here, we use the module template T with three proteins (A, B and C) and three PPIs (A–B, A–C and B–C) as an example to define the homologous modules of T as follows: (i) A′, B′ and C′ are the homologous proteins of A, B and C, respectively, with the significant sequence similarity (BLASTP E-values ≤ 10−10) (16); (ii) A′–B′, A′–C′ and B′–C′ are the homologous PPIs of A–B, A–C and B–C, respectively, with significant joint sequence similarity (joint E-value ≤ 10−40) (13); (iii) high topology similarity between modules A′–B′–C′ and A–B–C. The protein (or PPI) aligned ratio is defined as x/X, where x and X are the numbers of proteins (or PPIs) in the homologous module (e.g. A′–B–C′) and template (e.g. A–B–C), respectively. Here, the protein aligned ratio ≥ 0.5 and PPI aligned ratio ≥ 0.3 are considered as topology similarity between two modules according to the statistical analysis of 75 706 modules derived from 370 module templates in KEGG MODULE database with 1442 species (8) (Supplementary Figure S2). For each template in the MoNetFamily server (1975 non-redundant modules), we added its PPIs according to our previous sequence-based homologous PPIs (PPISearch with joint E-value ≤ 10−70) (13) and structure-based homologous PPIs (PCFamily with Z-value ≥ 4) (19) derived from the following PPI databases: (i) 461 077 experimental PPIs in the annotated PPI databases [IntAct (20), MIPS (21), DIP (22), MINT (23) and BioGRID (24)]; (ii) 9657 PPIs from PDB crystal structures (17). Please note that our analysis is limited to physical PPIs, specifically protein complexes.
MMI network
The MMI can be quantified by the PPIs between two modules (2,10). To determine the MMI between two modules, we added inter-module PPIs through our previous homologous PPIs derived from the six public PPI databases (13,19). Here, we decided the MMI based on the P-value of the hypergeometric distribution (10) defined as
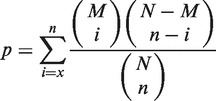 |
where x is observed annotated inter-module PPIs [e.g. x is 4 (red lines) between modules JAK2–PAFR–TYK2 and the hexameric human IL-6/IL-6α receptor/gp130 in Figure 2D]; i and n (e.g. n is 9 in Figure 2D) are the numbers of annotated inter-module PPIs and all combinational protein pairs between two modules, respectively; M and N are total numbers of annotated inter-module PPIs and all combinational protein pairs between any two modules in a MMI network, receptively. Here, the MMI between two modules should satisfy two criteria: (i) P-value ≤ 10−4 (the method to determine the threshold of P-value for MMIs was given in Supplementary Figures S10 and S11); (ii) at least two proteins of a module participate in inter-module PPIs. Finally, we identified 9678, 8942 and 8722 MMIs for the MMI networks of H. sapiens, M. musculus and D. rerio, respectively.
Annotations of query and modules
We annotated the query and modules by utilizing the consensus GO terms of homologous and neighboring modules. To annotate a module with Y proteins, we define a consensus ratio (CRM) of GO term i as CRM = Yi/Y, where Yi is the number of proteins with GO term i in a module. Next, the enrichment for each module in each GO term was determined by the P-value of the hypergeometric distribution and then this P-value was adjusted based on Bonferroni correction (25,26). Here, a GO term is considered as a representative GO term of a module if CRM > 0.6 and adjusted P-value of GO term ≤ 0.05 (25,26) based on statistically analysis. Furthermore, MoNetFamily used the consensus ratio of module family (CRF) and agreement ratio (AR), proposed by our previous works (13,19), to annotate the biological functions of the query proteins and a module family. The CRF is defined as CRF = Fa/F, where F is the total number of homologous modules in a module family; Fa is the number of homologous modules with representative GO term a in a module family. The AR is given as
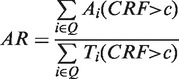 |
where Q is a set of query templates; Ti (CRF > c) is the total number of the representative GO terms of template i when CRF > c; Ai (CRF > c) is the number of the agreement representative GO terms of template i when CRF > c.
INPUT, OUTPUT and OPTIONS
The MoNetFamily is an easy-to-use web server (Supplementary Figure S1). Users input a set of protein sequence(s) in FASTA format, gene name(s), or UniProtKB accession number(s) (Figure 1B and Supplementary Figure S1A). Typically, the MoNetFamily server yields module candidates within 20 s when querying three sequences and the number of amino acids is less than 300 (Figures 1B). For a query, MoNetFamily shows details of hit module, MMIs (Figure 2D and Supplementary Figure S1C) and MMI networks using Cytoscape (27) across multiple species. For each module family, the server presents the homologous modules, the numbers of organisms and division groups, PPI profiles (Figure 1D and Supplementary Figure S1D) and conserved GO annotations (Figure 1C and Supplementary Figure S1B). In addition, users can download summarized results of query, module templates of query and module family and the neighboring modules across multiple species of the template.
MMI network analysis
We evaluated the properties and biological meanings of the MMI networks across multiple species (Figure 2A, B and Supplementary Figures S3–S6). Our derived MMI networks of H. sapiens, M. musculus and D. rerio were evaluated based on the characteristic of scale-free networks that the P(k), the probability of a node with k links, decreases as the node degree increases on a log–log plot (Figure 2B, Supplementary Figures S3A and S3B). The degree exponent γ are 1.183, 1.143 and 1.218 in the MMI networks of H. sapiens, M. musculus and D. rerio, respectively. This result is consistent with the architecture (i.e. weak scale-free network properties) of some cellular networks (28,29) and the MMI networks in S. cerevisiae (2,30) (Supplementary Figure S3C). A scale-free network typically has degree exponents 2 ≤ γ ≤ 3, but can also exist with γ < 2 (28,29). In addition, the median of degree (k) are 10, 10 and 11 for the networks of H. sapiens, M. musculus and D. rerio, respectively. The hubs, highly connected nodes, often play the key role in the network, such as the modules JAK2–PAFR–TYK2 (k = 25) in our derived MMI network.
Our derived MMI network can reflect the communication of five major cellular processes (Figure 2A), including nucleic acid metabolic process (e.g. transcription); protein metabolic process (e.g. translation); intracellular signal transduction process; integrin-mediated signal transduction process; and transport process. This MMI network presents the kernel processes (e.g. central dogma) performing the fundamental cellular metabolisms, that are transcription of nucleic acid metabolic process and translation of protein metabolic process. Signal transduction and transport processes, locating in cell membrane and cytoplasm, are the peripheral portion of the MMI network and communicate with two kernel processes (Figure 2A and Supplementary Figure S4).
Here, our derived MMI network was used to analyze the cell proliferation behavior via stimulations of the extracellular matrix (ECM) proteins and growth factors (Supplementary Figures S5 and S6). Cell proliferation is regulated by integrin-mediated adhesion to the ECM (e.g. ITGA5–ITGB1–FN1–TGM2 module in integrin-mediated signal transduction process) and the binding of growth factors to their receptors (e.g. JAK2–PAFR–TYK2 module in intracellular signal transduction process) (31). These two modules interact with Frs2–Grb2–Shp2 module (in intracellular signal transduction process) which transduces the signal to the ALL-1 supercomplex (in nucleic acid metabolic process) interacting with RNA polymerase II and TRAP–SMCC mediator modules. After transcription and translation, the newly synthesized proteins would be transported from endoplasmic reticulum to their destinations through the kinase maturation module 1 (in protein metabolic process) and the SNARE module (in transport process). In summary, MoNetFamily is a useful tool in the network biology to explore cellular processes. In the following two subsections, we utilized JAK2–PAFR–TYK2 module and TRAP–SMCC mediator module to describe MMIs and homologous modules.
Example analysis
JAK2–PAFR–TYK2 module
Module family of JAK2–PAFR–TYK2 module
Figure 1 shows the search results using Janus kinase 2 (Jak2, UniProt accession number: Q62120), platelet-activating factor receptor (Pafr or Ptafr, Q62035), and tyrosine kinase 2 (Tyk2, Q3U447) of M. musculus as the query (Figure 1B). For this query, the MoNetFamily server found the template candidate (JAK2–PAFR–TYK2 module, CORUM ID: 5178) (Figure 1B) and the homologous modules in 10 organisms, including H. sapiens, M. musculus, D. rerio and D4. Drosophila melanogaster (Figure 1D). JAK2–PAFR–TYK2 module, a mediator with diverse physiological and pathological actions, plays an important role for innate immune response in human skin (32,33). JAK2 and TYK2 are non-receptor tyrosine kinases of JAK family involving in mammalian development and immune disease (32,34). MoNetFamily annotated the JAK2–PAFR–TYK2 module family with GO terms (i.e. non-membrane spanning protein tyrosine kinase activity and cytoskeleton) and its neighbor modules with three GO terms, including epidermal growth factor receptor (EGFR) signaling pathway, signal transducer activity and cytosol (Figure 1C)
MMIs and neighboring modules
In the MMI network of H. sapiens, JAK2–PAFR–TYK2 module (red node) has 25 neighboring modules (green node). These 26 modules form a subnetwork and consist of 131 MMIs. (Figures 2A and 2C and Supplementary Table S1). According to GO term and MIPS FunCat (35) analysis, these modules can be roughly divided into three groups, cell surface receptor linked signaling pathway (orange), cellular protein metabolic pathway (purple) and interleukine receptor signaling pathway (blue). JAK2–PAFR–TYK2 module is a hub highly communicating with both intra-cellular and extra-cellular signal transduction processes (Figure 2C). Among these 25 neighboring modules, modules IL4–IL4R–IL2RG (CORUM ID: 1515) and RIN1–STAM2–EGFR (CORUM ID: 3678) are lack of homologous modules in D. rerio.
The module template, IL-6/IL-6Rα/gp130 (PDB code: 1p9m) from H. sapiens, participates in immunoregulatory mechanisms (36). According to our previous work, PCFamily (19), the binding model of IL-6/IL-6Rα/gp130 of H. sapiens is significantly different from the ones of M. musculus and D. rerio based on two observations: (i) For the interface between proteins gp130 and IL-6, the contact-residue (colored) identities of IL-6 between H. sapiens with M. musculus (7.7%) and D. rerio (0%) are very low (Supplementary Figure 7). (ii) The Z-values of interface similarities for the pair proteins gp130 and IL-6 are 0.923 (M. musculus) and −1.638 (D. rerio) using the structure template (PDB code: 1p9m) and the template-based scoring function (19).
Potential drug targets for psoriasis
Psoriasis is an autoimmune disease and one of the most common human skin diseases (37). Proteins JAK2 and TYK2 have been proposed as the potential targets for designing psoriasis drugs, such as ruxolitinib (38) and tasocitinib (39). Interestingly, the neighboring modules of JAK2–PAFR–TYK2 module derived by MoNetFamily can provide the clues for searching psoriasis target. Among 25 neighboring modules, 12 modules are highly conserved and annotated with cell surface receptor linked signaling pathway. Moreover, the module family annotations of SH3P2/OSTF1–CBL–SRC module, SLP-76–Cbl–Grb2–Shc module, Fc receptor gamma-R1 stimulated and CAS–SRC–FAK module belong to EGFR signaling pathway (Supplementary Table S1).
The inter-module PPIs between modules JAK2–PAFR–TYK2 and IL-6/IL6Rα/gp130 are JAK2–gp130, JAK2–IL-6Rα, TYK2–gp130 and TYK2–IL-6Rα (Figure 2D). Based on hypergeometric distribution, IL-6 was induced by JAK–STAT signal transduction pathway through the MMI (P-value = 1.06e − 5) between these two modules. According to increment of IL-6 levels after infection with HIV (40), the PPI JAK2–IL-6 should be considered as a potential target for controlling HIV-associated psoriasis. Additionally, JAK2–PAFR–TYK2 module simultaneously regulates EGFR and interleukine receptor signaling pathways. These observations imply the MMIs and networks of JAK2–PAFR–TYK2 and its neighboring modules provide a valuable insight for exploring the mechanisms of JAK-induced inflammatory diseases (e.g. psoriasis and rheumatoid arthritis).
TRAP–SMCC mediator module
TRAP–SMCC mediator module is the central regulator of the transcription apparatus (41,42). Mediator of RNA polymerase II transcription subunit 19 (Med19), the member of TRAP–SMCC mediator module, promotes tumorigenesis of lung (43) and breast (44) cancers. Using Med19 (Q8C1S0) of M. musculus as the query protein (Supplementary Figure S1), MoNetFamily found TRAP–SMCC mediator module and its module family with seven homologous modules. This family was annotated with three GO terms, including transcription (CRF = 0.83), RNA polymerase II transcription mediator activity (CRF = 1.00) and mediator complex (CRF = 1.00) (Supplementary Figure S1B). Interestingly, the GO annotations of its 15 neighboring modules (green node) in mammals (Supplementary Figure S1B) are highly consistent to the ones of TRAP–SMCC mediator module family. These results suggest that our server can utilize the GO annotations of the module family and its neighboring modules to predict the cellular functions of the query protein(s).
In the MMI networks of H. sapiens and M. musculus, TRAP–SMCC mediator module (red node) has 15 neighboring modules (green node) and these 16 modules form the subnetwork with 57 MMIs (Supplementary Figure S1B). According to GO terms and MIPS FunCat analysis, these modules dynamically regulate transcription and can be roughly divided into three groups, including transcription activation, transcription repression and DNA conformation modification (e.g. chromatin structure modification) (Supplementary Figure S8). Our MMIs shows that two proteins MED19 and mediator of RNA polymerase II transcription subunit 29 (IXL or MED29, B4DUA7) of TRAP–SMCC mediator module in mammals highly interact with its neighboring modules. These results imply that MED19 and MED29 play the key role in tumorigenesis and are consistent with the overexpression of these two proteins in breast, lung and pancreatic cancers (43,44).
Homologous module evaluation
To evaluate the accuracy of the MoNetFamily server for identifying homologous modules and the annotations of query protein(s), we selected a non-redundant template set, termed NRT with 1975 modules. The NRT set and the random module set are considered as the gold standard positive and negative sets, respectively. The random set consists of 98 750 (1975 × 50) modules by randomly generating 50 modules with the same number of proteins for each template in the NRT set. The consensus ratios (CRM) of GO terms (i.e. BP, CC and MF) in the module of NRT set are significantly higher than the ones of the random module set (Supplementary Figure S9). The CRM values (BP, CC and MF) of ∼70% (>1300) templates exceed 0.6 for NRT set; conversely, CRM values of the random set are 3.9% (BP), 8.1% (MF) and 18.3% (CC). For these GO terms with CRM > 0.6, the adjusted P-values of 88.2% (7776/8819) terms are < 0.05 for the NRT set.
Figure 3 shows the relationships between AR and CRF values of BP, CC and MF for homologous modules of the NRT set. If the CRF values of consensus GO terms (i.e. BP, CC and MF) of a module family are greater than 0.6, the AR values are consistently high for BP (0.68, green), CC (0.79, blue) and MF (0.79, red). For example, the representative GO terms (CRF > 0.6) of TRAP–SMCC mediator module family (seven homologous modules) are transcription (CRF = 0.83 and adjusted P-value = 4.59e − 08), RNA polymerase II transcription mediator activity (CRF = 1.00 and adjusted P-value = 1.41e − 11), and mediator complex (CRF = 1.00 and adjusted P-value = 1.42e − 05). These three representative GO terms can be used to annotate the module template, TRAP–SMCC mediator module. These results indicate that MoNetFamily achieves high agreements on consensus GO terms between the queries (i.e. module templates) and their respective homologous modules. Furthermore, the module templates and their homologous modules derived by our method often possess specific biological functions.
Figure 3.
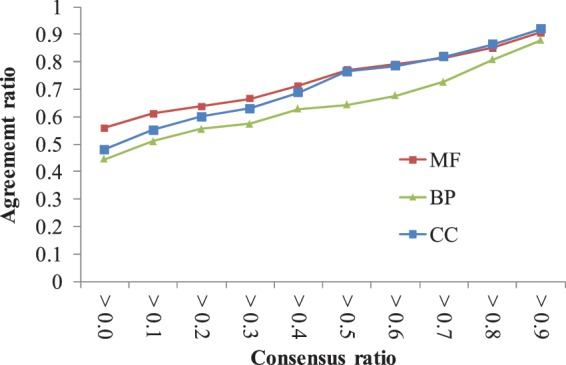
Evaluation module annotations on the NRT set. The relationships between agreement ratios (AR) and the consensus ratios (CRF) of BP, MF and CC using 1975 module families.
CONCLUSIONS
This work demonstrates the utility and feasibility of using the MoNetFamily server to identify MMIs and MMI networks in vertebrates through homologous modules. MoNetFamily is the first server to provide the neighboring modules and MMIs in module networks across multiple species; the profiles of proteins and PPIs in module families; GO annotations of neighboring modules and module families. Our results indicate that the server can be useful for MMI network visualizations across multiple vertebrates and annotating a set of query proteins by using module families and neighboring modules. We believe that MoNetFamily is a fast homologous modules and MMIs search server and is able to provide valuable insights for studying the module evolution and cellular processes.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table 1, Supplementary Figures 1–11.
FUNDING
Funding for open access charge: National Science Council, partial supports of Ministry of Education and National Health Research Institutes [NHRI-EX100-10009PI]; ‘Center for Bioinformatics Research of Aiming for the Top University Program’ of the National Chiao Tung University and Ministry of Education, Taiwan. J.-M. Yang also thanks Core Facility for Protein Structural Analysis supported by National Core Facility Program for Biotechnology.
Conflict of interest statement. None declared.
REFERENCES
- 1.Malovannaya A, Lanz RB, Jung SY, Bulynko Y, Le NT, Chan DW, Ding C, Shi Y, Yucer N, Krenciute G, et al. Analysis of the human endogenous coregulator complexome. Cell. 2011;145:787–799. doi: 10.1016/j.cell.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang H, Kakaradov B, Collins SR, Karotki L, Fiedler D, Shales M, Shokat KM, Walther TC, Krogan NJ, Koller D. A complex-based reconstruction of the Saccharomyces cerevisiae interactome. Mol. Cell. Proteomics. 2009;8:1361–1381. doi: 10.1074/mcp.M800490-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics. 2003;4:2. doi: 10.1186/1471-2105-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Segal E, Shapira M, Regev A, Pe'er D, Botstein D, Koller D, Friedman N. Module networks: identifying regulatory modules and their condition-specific regulators from gene expression data. Nat. Genet. 2003;34:166–176. doi: 10.1038/ng1165. [DOI] [PubMed] [Google Scholar]
- 5.Snel B, Huynen MA. Quantifying modularity in the evolution of biomolecular systems. Genome Res. 2004;14:391–397. doi: 10.1101/gr.1969504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campillos M, von Mering C, Jensen LJ, Bork P. Identification and analysis of evolutionarily cohesive functional modules in protein networks. Genome Res. 2006;16:374–382. doi: 10.1101/gr.4336406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jensen LJ, Kuhn M, Stark M, Chaffron S, Creevey C, Muller J, Doerks T, Julien P, Roth A, Simonovic M, et al. STRING 8–a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009;37:D412–D416. doi: 10.1093/nar/gkn760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T, et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36:D480–D484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ruepp A, Waegele B, Lechner M, Brauner B, Dunger-Kaltenbach I, Fobo G, Frishman G, Montrone C, Mewes HW. CORUM: the comprehensive resource of mammalian protein complexes–2009. Nucleic Acids Res. 2010;38:D497–D501. doi: 10.1093/nar/gkp914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bandyopadhyay S, Mehta M, Kuo D, Sung MK, Chuang R, Jaehnig EJ, Bodenmiller B, Licon K, Copeland W, Shales M, et al. Rewiring of genetic networks in response to DNA damage. Science. 2010;330:1385–1389. doi: 10.1126/science.1195618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, Pang N, Forslund K, Ceric G, Clements J, et al. The Pfam protein families database. Nucleic Acids Res. 2012;40:D290–D301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Andreeva A, Howorth D, Brenner SE, Hubbard TJ, Chothia C, Murzin AG. SCOP database in 2004: refinements integrate structure and sequence family data. Nucleic Acids Res. 2004;32:D226–D229. doi: 10.1093/nar/gkh039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen CC, Lin CY, Lo YS, Yang JM. PPISearch: a web server for searching homologous protein-protein interactions across multiple species. Nucleic Acids Res. 2009;37:W369–W375. doi: 10.1093/nar/gkp309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kersey P, Bower L, Morris L, Horne A, Petryszak R, Kanz C, Kanapin A, Das U, Michoud K, Phan I, et al. Integr8 and genome reviews: integrated views of complete genomes and proteomes. Nucleic Acids Res. 2005;33:D297–D302. doi: 10.1093/nar/gki039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu H, Luscombe NM, Lu HX, Zhu X, Xia Y, Han JD, Bertin N, Chung S, Vidal M, Gerstein M. Annotation transfer between genomes: protein-protein interologs and protein-DNA regulogs. Genome Res. 2004;14:1107–1118. doi: 10.1101/gr.1774904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen YC, Lo YS, Hsu WC, Yang JM. 3D-partner: a web server to infer interacting partners and binding models. Nucleic Acids Res. 2007;35:W561–W567. doi: 10.1093/nar/gkm346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lo YS, Lin CY, Yang JM. PCFamily: a web server for searching homologous protein complexes. Nucleic Acids Res. 2010;38:W516–W522. doi: 10.1093/nar/gkq464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aranda B, Achuthan P, Alam-Faruque Y, Armean I, Bridge A, Derow C, Feuermann M, Ghanbarian AT, Kerrien S, Khadake J, et al. The IntAct molecular interaction database in 2010. Nucleic Acids Res. 2010;38:D525–D531. doi: 10.1093/nar/gkp878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mewes HW, Dietmann S, Frishman D, Gregory R, Mannhaupt G, Mayer KF, Munsterkotter M, Ruepp A, Spannagl M, Stumpflen V, et al. MIPS: analysis and annotation of genome information in 2007. Nucleic Acids Res. 2008;36:D196–D201. doi: 10.1093/nar/gkm980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xenarios I, Salwinski L, Duan XJ, Higney P, Kim SM, Eisenberg D. DIP, the Database of Interacting Proteins: a research tool for studying cellular networks of protein interactions. Nucleic Acids Res. 2002;30:303–305. doi: 10.1093/nar/30.1.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ceol A, Chatr Aryamontri A, Licata L, Peluso D, Briganti L, Perfetto L, Castagnoli L, Cesareni G. MINT, the molecular interaction database: 2009 update. Nucleic Acids Res. 2010;38:D532–D539. doi: 10.1093/nar/gkp983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stark C, Breitkreutz BJ, Chatr-Aryamontri A, Boucher L, Oughtred R, Livstone MS, Nixon J, Van Auken K, Wang X, Shi X, et al. The BioGRID interaction database: 2011 update. Nucleic Acids Res. 2011;39:D698–D704. doi: 10.1093/nar/gkq1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Medina I, Carbonell J, Pulido L, Madeira SC, Goetz S, Conesa A, Tarraga J, Pascual-Montano A, Nogales-Cadenas R, Santoyo J, et al. Babelomics: an integrative platform for the analysis of transcriptomics, proteomics and genomic data with advanced functional profiling. Nucleic Acids Res. 2010;38:W210–W213. doi: 10.1093/nar/gkq388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boyle EI, Weng S, Gollub J, Jin H, Botstein D, Cherry JM, Sherlock G. GO::TermFinder–open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics. 2004;20:3710–3715. doi: 10.1093/bioinformatics/bth456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lopes CT, Franz M, Kazi F, Donaldson SL, Morris Q, Bader GD. Cytoscape Web: an interactive web-based network browser. Bioinformatics. 2010;26:2347–2348. doi: 10.1093/bioinformatics/btq430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barabasi AL, Oltvai ZN. Network biology: understanding the cell's functional organization. Nat. Rev. Genet. 2004;5:101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- 29.Seyed-Allaei H, Bianconi G, Marsili M. Scale-free networks with an exponent less than two. Phys. Rev. E, Stat. Nonlinear Soft Matter Phys. 2006;73:046113. doi: 10.1103/PhysRevE.73.046113. [DOI] [PubMed] [Google Scholar]
- 30.Li SS, Xu K, Wilkins MR. Visualization and analysis of the complexome network of Saccharomyces cerevisiae. J. Proteome Res. 2011;10:4744–4756. doi: 10.1021/pr200548c. [DOI] [PubMed] [Google Scholar]
- 31.Schwartz MA, Assoian RK. Integrins and cell proliferation: regulation of cyclin-dependent kinases via cytoplasmic signaling pathways. J. Cell Sci. 2001;114:2553–2560. doi: 10.1242/jcs.114.14.2553. [DOI] [PubMed] [Google Scholar]
- 32.Lukashova V, Chen Z, Duhe RJ, Rola-Pleszczynski M, Stankova J. Janus kinase 2 activation by the platelet-activating factor receptor (PAFR): roles of Tyk2 and PAFR C terminus. J. Immunol. (Baltimore, Md.: 1950) 2003;171:3794–3800. doi: 10.4049/jimmunol.171.7.3794. [DOI] [PubMed] [Google Scholar]
- 33.Fridman JS, Scherle PA, Collins R, Burn T, Neilan CL, Hertel D, Contel N, Haley P, Thomas B, Shi J, et al. Preclinical evaluation of local JAK1 and JAK2 inhibition in cutaneous inflammation. J. Investigative Dermatol. 2011;131:1838–1844. doi: 10.1038/jid.2011.140. [DOI] [PubMed] [Google Scholar]
- 34.Gu J, Wang Y, Gu X. Evolutionary analysis for functional divergence of Jak protein kinase domains and tissue-specific genes. J. Mol. Evol. 2002;54:725–733. doi: 10.1007/s00239-001-0072-3. [DOI] [PubMed] [Google Scholar]
- 35.Ruepp A, Zollner A, Maier D, Albermann K, Hani J, Mokrejs M, Tetko I, Guldener U, Mannhaupt G, Munsterkotter M, et al. The FunCat, a functional annotation scheme for systematic classification of proteins from whole genomes. Nucleic Acids Res. 2004;32:5539–5545. doi: 10.1093/nar/gkh894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boulanger MJ, Chow DC, Brevnova EE, Garcia KC. Hexameric structure and assembly of the interleukin-6/IL-6 alpha-receptor/gp130 complex. Science. 2003;300:2101–2104. doi: 10.1126/science.1083901. [DOI] [PubMed] [Google Scholar]
- 37.Lowes MA, Bowcock AM, Krueger JG. Pathogenesis and therapy of psoriasis. Nature. 2007;445:866–873. doi: 10.1038/nature05663. [DOI] [PubMed] [Google Scholar]
- 38.Mesa RA. Ruxolitinib, a selective JAK1 and JAK2 inhibitor for the treatment of myeloproliferative neoplasms and psoriasis. IDrugs Investigational Drugs J. 2010;13:394–403. [PubMed] [Google Scholar]
- 39.Chrencik JE, Patny A, Leung IK, Korniski B, Emmons TL, Hall T, Weinberg RA, Gormley JA, Williams JM, Day JE, et al. Structural and thermodynamic characterization of the TYK2 and JAK3 kinase domains in complex with CP-690550 and CMP-6. J. Mol. Biol. 2010;400:413–433. doi: 10.1016/j.jmb.2010.05.020. [DOI] [PubMed] [Google Scholar]
- 40.Poli G, Bressler P, Kinter A, Duh E, Timmer WC, Rabson A, Justement JS, Stanley S, Fauci AS. Interleukin 6 induces human immunodeficiency virus expression in infected monocytic cells alone and in synergy with tumor necrosis factor alpha by transcriptional and post-transcriptional mechanisms. J. Exp. Med. 1990;172:151–158. doi: 10.1084/jem.172.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Malik S, Wallberg AE, Kang YK, Roeder RG. TRAP/SMCC/mediator-dependent transcriptional activation from DNA and chromatin templates by orphan nuclear receptor hepatocyte nuclear factor 4. Mol. Cell. Biol. 2002;22:5626–5637. doi: 10.1128/MCB.22.15.5626-5637.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Malik S, Roeder RG. Dynamic regulation of pol II transcription by the mammalian Mediator complex. Trends Biochem Sci. 2005;30:256–263. doi: 10.1016/j.tibs.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 43.Sun M, Jiang R, Li JD, Luo SL, Gao HW, Jin CY, Shi DL, Wang CG, Wang B, Zhang XY. MED19 promotes proliferation and tumorigenesis of lung cancer. Mol. Cell. Biochem. 2011;355:27–33. doi: 10.1007/s11010-011-0835-0. [DOI] [PubMed] [Google Scholar]
- 44.Kuuselo R, Savinainen K, Sandstrom S, Autio R, Kallioniemi A. MED29, a component of the mediator complex, possesses both oncogenic and tumor suppressive characteristics in pancreatic cancer. Int. J. cancer. J. Int du Cancer. 2011;129:2553–2565. doi: 10.1002/ijc.25924. [DOI] [PMC free article] [PubMed] [Google Scholar]