Abstract
Evaluating the specificity of polymerase chain reaction (PCR) primers is an essential step in PCR primer design. The MFEprimer-2.0 server allows users to check primer specificity against genomic DNA and messenger RNA/complementary DNA sequence databases quickly and easily. MFEprimer-2.0 uses a k-mer index algorithm to accelerate the search process for primer binding sites and uses thermodynamics to evaluate binding stability between each primer and its DNA template. Several important characteristics, such as the sequence, melting temperature and size of each amplicon, either specific or non-specific, are reported on the results page. Based on these characteristics and the user-friendly output, users can readily draw conclusions about the specificity of PCR primers. Analyses for degenerate primers and multiple PCR primers are also supported in MFEprimer-2.0. In addition, the databases supported by MFEprimer-2.0 are comprehensive, and custom databases can also be supported on request. The MFEprimer-2.0 server does not require a login and is freely available at http://biocompute.bmi.ac.cn/CZlab/MFEprimer-2.0. More over, the MFEprimer-2.0 command-line version and local server version are open source and can be downloaded at https://github.com/quwubin/MFEprimer/wiki/Manual/.
INTRODUCTION
Checking the specificity of polymerase chain reaction (PCR) primers is a key step in primer design. Several primer design programs such as PerlPrimer and Primer3Plus suggest using National Center for Biotechnology Information's Basic Local Alignment Search Tool (NCBI BLAST) (1) to examine primer specificity. Other primer specificity-checking programs such as virtual PCR (2), PRImer Match EXtractor (3), Primer-UniGene Selectivity (4) and GenomeTester (5) also focus on sequence similarity analyses between the primers and DNA templates, although the importance of other factors in running a successful PCR reaction is well documented. These factors have been described in detail in our previous work (6) and include the stability of the 3′ end of the primer and its melting temperature (Tm). Hybridisation between the PCR primer and the DNA template is a thermodynamic reaction (7). Thus, it is not sufficient to determine binding sites by merely using sequence alignment programs such as BLAST. For example, the G–C and A–T matches have equal scores in an NCBI BLAST search, but the G–C match is more stable than the A–T match. Even a mismatch, such as G–G, contributes as much as -2.2 kcal/mol (Gibbs free energy) to the duplex stability (8). Therefore, the thermodynamic approach should be applied for reasonable predictions of unintended primer binding sites. In addition, the majority of the current PCR primer analysis programs require long running times when checking the specificity of PCR primers against large genomic DNA (gDNA) databases.
Here, we introduce MFEprimer-2.0, a fast and thermodynamics-based PCR primer specificity-checking program, representing a significant update of our previous work, MFEprimer (6). New features in MFEprimer-2.0 include the following: (i) the use of the nearest-neighbour (NN) model (9) rather than the NCBI BLAST program to evaluate binding stabilities between a primer and its binding sites; (ii) a k-mer index algorithm to significantly accelerate the binding-site search process; (iii) a redesigned homepage and results page to improve the user experience; (iv) support for degenerate primer analysis; (v) support for multiple databases for cross-species PCR primer assays and (vi) post-analysis of the predicted amplicons, for example, the “MultiAlign” function, to check for similarities among the predicted amplicons.
THE MFEPRIMER-2.0 ALGORITHM
A successful PCR reaction typically requires stable and specific binding between the 3′ ends of the primer and its DNA template; in contrast, the 5′ end is not critical for PCR. The uniqueness of the 3′-end subsequence of a primer mainly determines the specificity of the primer, although the binding stability is related to the entire primer sequence. The main MFEprimer-2.0 evaluation process consists of four steps: (i) find all of the binding-site positions of the 3′-end subsequence of a primer among all possible DNA template sequences, including competing sequences; (ii) evaluate the binding stability of the entire primer sequence using the NN model; (iii) run a virtual PCR amplification and (iv) filter out the predicted amplicons by size and other parameters.
Determining the maximum size of the 3′-end subsequence
The annealing temperature of a successful PCR reaction is usually more than 55°C to prevent non-specific PCR amplification. Therefore, primers with Tm values less than 55°C will not perform properly in PCR reactions. We must thus determine the maximum size of the 3′-end subsequence such that the Tm value of the most stable subsequence is less than 55°C. In MFEprimer-2.0, to avoid missing any stable binding sites, a stricter cut-off value of 48°C was set to determine the maximum size of the 3′-end subsequence. A Python (http://www.python.org) script (see Supplementary Scripts) was used to calculate all of the k-mer values, with k ranging from 5 to 9. Here, a k-mer is defined as a short DNA sequence with a length of k nucleotides. The results revealed that k = 9 is suitable for the maximum size of the 3′-end subsequence (see Supplementary Data).
The k-mer index algorithm
The k-mer index algorithm is similar to the “database formatting” process in sequence similarity searching programs, such as BLAST (1) or BLAT (10). The basic idea of the k-mer index algorithm is the following: first, all the positions of all k-mers are stored in a relational database; second, for a given primer, its 3′-end subsequence (here, a k-mer) is constant, and we only need to retrieve the position information from the relational database. In MFEprimer-2.0, we use k = 9 and a sliding window size of 1 bp to index the gDNA and complementary DNA (cDNA) databases stored in our server. MFEprimer-2.0 now supports the most widely used databases such as human gDNA sequences and that of other model organisms and cDNA databases. The mitochondrial DNA sequence for each species is also included in the corresponding gDNA database. Custom databases can be supported upon request and indexed for PCR primer evaluation.
Evaluating the binding stability of the entire primer sequence using the NN model
For each of the binding sites, MFEprimer-2.0 first retrieves the subsequence of the potential DNA template with the same length as the primer, based on the position of the binding site. Secondly, MFEprimer-2.0 uses the NN model to calculate the duplex stability formed by the subsequence and the primer sequence. Only thermodynamically stable mismatches (8) are allowed in the binding pattern. The Tm value and Gibbs free energy (ΔG) for each binding pattern are calculated and shown on the results page.
THE MFEPRIMER-2.0 SERVER
The MFEprimer-2.0 server is available at http://biocompute.bmi.ac.cn/CZlab/MFEprimer-2.0. The web site is freely accessible and does not require a login.
Inputs
There are two mandatory inputs in MFEprimer-2.0: primer sequence and a database selection. A “Batch Mode” is available for the batch evaluation of primer sequences. This mode also supports multiple database selections for cross-species PCR primer analyses. Other parameters such as “Results filter settings” have default values for routine analyses.
Running time
When choosing “Single Mode”, the running time of MFEprimer-2.0 is usually 1–10 seconds because the database selection is limited to one species. However, the same task in MFEprimer (Version 1.0) requires several minutes to complete. In “Batch Mode”, the running time of MFEprimer-2.0 is unpredictable, and a job queue control system is used to manage these tasks when multiple databases and/or multiple primer sequences are selected. Users can refresh the browser several minutes later to check the status of the job. The results are kept on the server for 24 hours.
Outputs
The MFEprimer-2.0 result page comprises five sections: (i) Query: the list of user input primer sequences with size, GC content and Tm value annotation; (ii) Brief descriptions of all the potential amplicons predicted by MFEprimer-2.0; (iii) Amplicon details: information on hybridisation details for each predicted amplicon and the amplicon sequence; (iv) Parameters: the parameters used during the evaluation process; (v) Citation: the first study reporting the MFEprimer program. Detailed information for the output is provided in the FAQ list (http://code.google.com/p/mfeprimer/wiki/FAQ).
Example analysis
Suppose that we have designed a pair of primers for amplification of the two splicing variants (GenBank No. NM_010923 and NM_180960) of the mouse Nnat (GeneID: 18111) gene. Before performing the PCR experiment in the laboratory, we can check the specificity of the primers and determine which amplicons are likely to be amplified. Two primers were designed with VizPrimer (11): a forward primer, GACCAGTAGACCTCGGCGAA, and a reverse primer, ACCTTGGCAAGTGCTCCTCT. These two primers were input into MFEprimer-2.0, and the database “Mouse–RNA & Genomic” was selected for specificity checking. The default values were used for the other parameters.
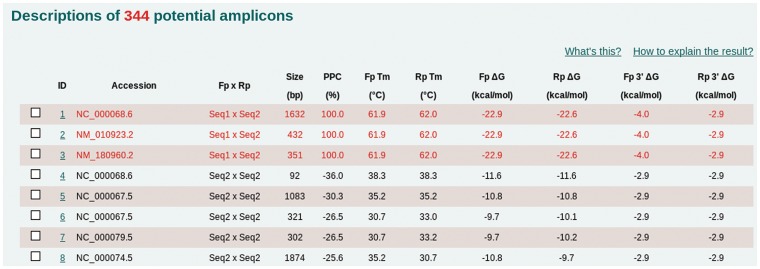
Figure 1 shows the “Description of x potential amplicons” section of the results page. Except for the amplicons marked with red boxes, the other amplicons have lower binding stability, as indicated by lower Tm and ΔG values. These results indicate that these amplicons would likely not exist under real PCR conditions, which are optimal for the target amplicon (here, the Tm is 61.9°C). On the contrary, the amplicons marked with the red boxes have the same binding stability and would be amplified in a real PCR experiment. These two sequences, NM_010923 (ID: 2) and NM_180960 (ID: 3), are the two splicing variants of the mouse Nnat gene and are the target amplicons.
Figure 1.
An example result using MFEprimer-2.0 for a primer pair designed for the mouse Nnat gene revealed many potential non-specific amplicons. Amplicons marked with red boxes have higher binding stability (Tm and ΔG values) than the others. The GenBank sequences NM_010923 (ID: 2) and NM_180960 (ID: 3) are the two splice variants of the Nnat gene, whereas NC_000068 (ID: 1) is the corresponding gDNA sequence region.
Here, we aimed to answer the question: is the other amplicon (ID: 1) a non-specific amplicon? The “MultiAlign” function can help the user identify the relationship between these amplicons (for detailed information, see https://github.com/quwubin/MFEprimer/wiki/HowTo). This analysis revealed that the NC_000068 (ID: 1) sequence is the gDNA sequence region of the mouse Nnat gene. In this case, the specificity of the primer pair was determined to be sufficient when the annealing temperature was optimal for the target amplicon (Tm = 61.9°C). In addition, these results indicated that if mouse gDNA was selected as the DNA template source, this primer pair would create an amplicon of 1632 bp. We experimentally validated the primer pair and obtained high-quality results (see https://github.com/quwubin/MFEprimer/wiki/HowTo for detailed results).
CONCLUSIONS
The k-mer index algorithm and the NN model increase the speed of MFEprimer-2.0 while preventing the omission of any possible and thermodynamically stable binding sites. With MFEprimer-2.0, users can now check the specificity of primers against entire gDNA databases in a few seconds and even against the combination of gDNA and cDNA/mRNA databases. In future, we will continue to update MFEprimer-2.0 to support SNP-based genotype assays and multiplex PCR primer analyses.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Data and Supplementary Scripts.
FUNDING
National Basic Research Project (973 program) [2012CB518200]; General Program [30900862, 30973107, 81070741, 81172770] of the Natural Science Foundation of China; State Key Laboratory of Proteomics of China [SKLP-O201104, SKLP-K201004, SKLP-O201002]; and Special Key Programs for Science and Technology of China [2012ZX09102301-016]. Funding for open access charge: the State Key Laboratory of Proteomics of China [SKLP-O201104, SKLP-K201004].
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We are grateful to the users who provided valuable suggestions during the development of MFEprimer-2.0. Some of them are listed on our “Thanks” page: https://github.com/quwubin/MFEprimer/wiki/iThank.
REFERENCES
- 1.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic. Acids. Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lexa M, Horak J, Brzobohaty B. Virtual PCR. Bioinformatics. 2001;17:192–193. doi: 10.1093/bioinformatics/17.2.192. [DOI] [PubMed] [Google Scholar]
- 3.Lexa M, Valle G. PRIMEX: rapid identification of oligonucleotide matches in whole genomes. Bioinformatics. 2003;19:2486–2488. doi: 10.1093/bioinformatics/btg350. [DOI] [PubMed] [Google Scholar]
- 4.Boutros PC, Okey AB. PUNS: transcriptomic- and genomic-in silico PCR for enhanced primer design. Bioinformatics. 2004;20:2399–2400. doi: 10.1093/bioinformatics/bth257. [DOI] [PubMed] [Google Scholar]
- 5.Andreson R, Reppo E, Kaplinski L, Remm M. GENOMEMASKER package for designing unique genomic PCR primers. BMC Bioinformatics. 2006;7:172. doi: 10.1186/1471-2105-7-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qu W, Shen Z, Zhao D, Yang Y, Zhang C. MFEprimer: multiple factor evaluation of the specificity of PCR primers. Bioinformatics. 2009;25:276–278. doi: 10.1093/bioinformatics/btn614. [DOI] [PubMed] [Google Scholar]
- 7.SantaLucia J., Jr Physical principles and visual-OMP software for optimal PCR design. Methods. Mol. Biol. 2007; 402:3–34. doi: 10.1007/978-1-59745-528-2_1. [DOI] [PubMed] [Google Scholar]
- 8.SantaLucia J, Hicks D. The thermodynamics of DNA structural motifs. Annu. Rev. Biophys. Biomol. Struct. 2004;33:415–440. doi: 10.1146/annurev.biophys.32.110601.141800. [DOI] [PubMed] [Google Scholar]
- 9.SantaLucia J. A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor hermodynamics. Proc. Natl. Acad. Sci. U.S.A. 1998;95:1460–1465. doi: 10.1073/pnas.95.4.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kent WJ. BLAT–the BLAST-like alignment tool. Genome. Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou Y, Qu W, Lu Y, Zhang Y, Wang X, Zhao D, Yang Y, Zhang C. VizPrimer: a web server for visualized PCR primer design based on known gene structure. Bioinformatics. 2011;27:3432–3434. doi: 10.1093/bioinformatics/btr582. [DOI] [PubMed] [Google Scholar]