Abstract
A major challenge in the post-genomic era is a better understanding of how human genetic alterations involved in disease affect the gene products. The KD4v (Comprehensible Knowledge Discovery System for Missense Variant) server allows to characterize and predict the phenotypic effects (deleterious/neutral) of missense variants. The server provides a set of rules learned by Induction Logic Programming (ILP) on a set of missense variants described by conservation, physico-chemical, functional and 3D structure predicates. These rules are interpretable by non-expert humans and are used to accurately predict the deleterious/neutral status of an unknown mutation. The web server is available at http://decrypthon.igbmc.fr/kd4v.
INTRODUCTION
A wide variety of human diseases have been linked to non-synonymous SNPs (nsSNPs), also called Missense Variants, which result in an alteration of the amino acid sequence of the encoded protein and can affect the function, solubility or structure of the mutated protein. Today, with the huge amount of protein information available in various biomedical databases, it is now possible to better understand the correlation between a nsSNP and the associated phenotypes.
Several methods (1) have been developed to predict the effects of nsSNPs on the 3D structure of a protein and its function, based on the hypothesis that variants that modify the structure/function at the molecular level are more likely to be deleterious. The methods can be divided into two main categories: (i) sequence-based methods using multiple sequence alignments and incorporating different approaches to quantify residue conservation: SIFT (2), PANTHER (3), SNAP(4) and SNP/GO (5) and (ii) methods combining sequence and 3D structure features such as the widely used Polyphen-2 (6), nsSNPAnalyzer (7) and SNPs3D (8). Most of these methods can classify a nsSNP as either deleterious (strong functional effect) or neutral (weak functional effect) with high accuracy. However, they only provide a final score and in general, no information is provided that could be used to evaluate the classification and to estimate the relationships between genotypic and phenotypic variation.
To overcome these limitations, the KD4v (Comprehensible Knowledge Discovery System for Missense Variant) server aims to discover, exploit and provide the user with links between the computed impact of a mutation and the human disease phenotype. We applied the ILP method (9) to a set of nsSNPs involved in human diseases that are mapped to 3D structure and annotated by the MSV3d (MisSense Variant mapped to 3D structure) pipeline (10). KD4v provides two complementary services: (i) a knowledgebase consisting of ILP rules based on 16 sequence/structure/evolution predicates that characterize deleterious mutations in any human gene and that can be interpreted by biologists and (ii) a tool for mutation prediction based on the ILP rules with performances similar to the most widely used methods: PolyPhen-2 and SIFT. In addition, the KD4v server links the human genes to a rich set of up-to-date information encompassing tissue expression, protein-protein interactions or phenotypic descriptions hosted by SM2PH (11).
MATERIALS AND METHODS
Missense variant annotation
The nsSNPs observed in all human proteins were annotated by the MSV3d pipeline, which automatically performs a sequence/structure/evolution analysis and has been shown to be robust and efficient (6,12). This includes various parameters which describe, among others, the physico-chemical changes induced by the amino acid substitution, the conservation pattern of the mutated residue, the status of mutated residues with respect to functional features. In KD4v, this multi-level sequence-based characterization of nsSNPs is complemented by parameters related to 3D models or the 3D Fold classification in SCOP (13). This results in pre-computed annotations for over 63 000 known nsSNPs in the 10 713 proteins with known or modelled 3D structures currently available. In addition, the user can also request a prediction for any new or unknown missense variant, if the protein can be mapped to a 3D structure.
The characterization of the background conservation and exploitation of the different types of evolutionary data has been described in detail previously (10). Briefly, we used MACSIMS (14) to annotate a multiple alignment, containing both Uniprot and PDB sequences, with information such as: (i) taxonomic data, (ii) functional descriptions, (iii) known domains or domains similar to a known 3D structure, (iv) potential disordered regions, (v) blocks that do not correspond to disordered regions or known domains but that are conserved at the family or subfamily level and thus may constitute uncharacterized domains and (vi) conservation pattern of domains and residues. If the variant position is mapped to an identified 3D structure, the structural context of each individual mutation is modelled based on several descriptors combining sequence/structure-related data using several software tools such as MODELLER (15), CSU (16), I-Mutant (17). Details of the predicates used in the KD4v server and computational methods/software are provided on the KD4v help page.
Dataset compilation and computer resource
We used the variant set from the Polyphen-2 training set (6) extracted from SwissVar (18) to train and test the KD4v server. Only nsSNPs that are mapped to 3D structures were retained and randomly split into a training set (6000 disease-causing mutations associated with distinct 881 OMIM phenotypes and 2000 neutral polymorphisms) and a first validation set (658 disease-causing variants associated with 311 distinct OMIM phenotypes and 298 neutral polymorphisms). We also created a second validation set (173 disease-causing mutations associated with distinct 39 OMIM phenotypes and 179 neutral polymorphisms), in which not only variants, but also protein sequences, were different in the training and validation sets. Our goal is to predict the deleterious nature of human variants, i.e. those variants associated with disease phenotypes, and it should be noted that these datasets do not specifically identify mutations that have a weaker effect on the function of the protein. The datasets are available for download from our website.
To guarantee a permanent powerful CPU resource for the KD4v server, we deployed the software on the Décrypthon grid (19) including a total of 58 machines and 475 processors under the AIX operating system distributed on six nodes.
Induction logic programming implementation
Induction Logic Programming (ILP) combines Machine Learning and Logic Programming (9). Briefly, given a formal encoding of the background knowledge and a set of examples, an ILP system will derive hypotheses explaining all positive examples and none, or almost none, negative examples. In this approach, logic is used as a language to induce hypotheses from the examples and background knowledge. The result of the learning step is a set of rules represented as logical formula, typically a Prolog program, that can be reused as a prediction service. The creation of the KD4v is based on distinct predicates deduced from the multi-level characterization provided by MSV3d (Supplementary Table S1) and involves various steps detailed in Supplementary Figure S1. We have limited our study to the task of discriminating the mutations linked to human diseases (deleterious) from those associated with the ‘polymorphism’ term (neutral). Thus, a positive example in Prolog syntax is defined as: ‘is_deleterious(m_Q92947.p.Gly390Ala)’ which indicates that, in protein Q92947, the replacement of the glycine at position 390 by an alanine is deleterious.
The implementation of the server also includes the optimization of the predicates using a 5-fold cross-validation on the training set with standard performance indicators including sensitivity, specificity, precision, recall, accuracy and F-measure (see legend of Supplementary Table S2 for a complete description). Thus, the final ILP model consists of 16 predicates (Supplementary Table S1) which can be separated into two major types: predicates describing the mutated residue or protein (functional and structural features) and predicates describing the physical, chemical or structural changes introduced by the substitution.
KD4v RULE SERVICE
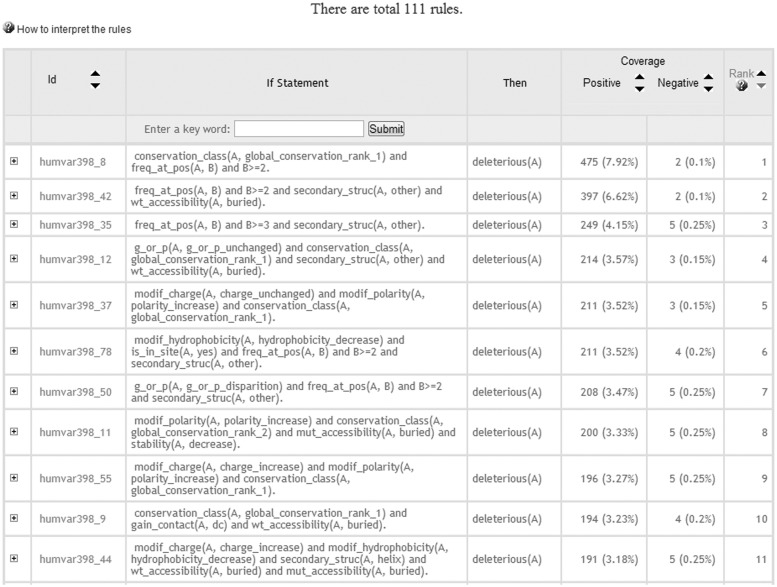
Currently, the server hosts 111 rules that are comprehensible by humans. These ILP rules can be used, for example, to uncover the relationships between the deleterious effect of a mutation and the multi-class conservation pattern or the type of the physico-chemical alterations (e.g. size, charge and hydrophobicity) introduced by the substitution. Figure 1 shows some induced rules on the web page. To illustrate how to interpret ILP rules, we can consider the humvar398_44 rule:
deleterious(A) :-
modif_charge(A, charge_increase) and
modif_hydrophobicity(A, hydrophobicity_decrease) and
secondary_struc(A, helix) and wt_accessibility(A, buried) and
mut_accessibility(A, buried).
Figure 1.
ILP rules. The first column provides a link to the positive (deleterious mutations) and negative (neutral mutations) examples covered by a given rule and that can be seen by clicking on the + icon. The second column provides the rule identifier (Id). The next two columns provide the ‘if’ and ‘then’ clauses of the induced rules. The two right most columns indicate the number of positive and negative examples covered by the rule in each row.
This rule states that a mutation A is deleterious if: (i) the charge of the residue is increased by the mutation; (ii) its hydrophobicity is decreased; (iii) the residue is found in a helix; (iv) the wild-type residue is buried; and (v) the mutant residue is also buried. This rule correctly identified 191 (3.18% of the 6000 studied) deleterious mutations, while misclassifying five neutral mutations as deleterious (0.25% of the 2000 neutral mutations in the training set).
KD4v PREDICTION SERVICE
Input and output
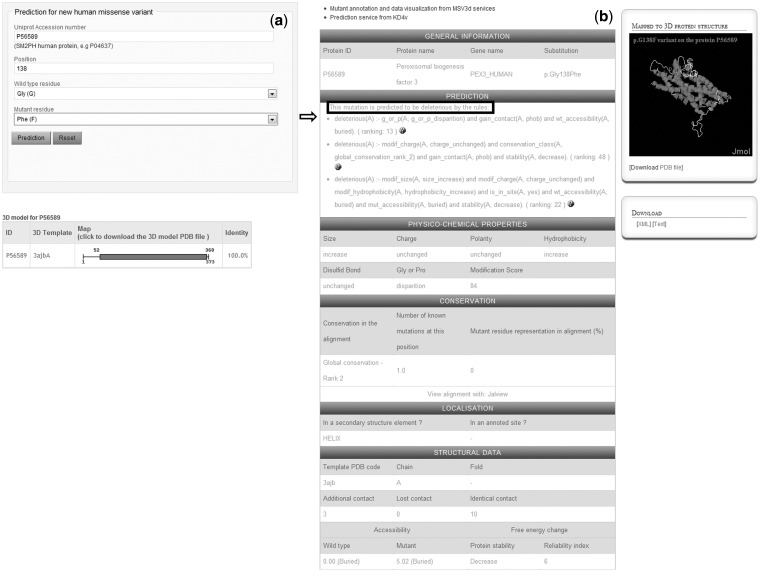
KD4v provides a service aimed at estimating nsSNP effects based on the ILP rules. It can be accessed via the web interface or via the SOAP Web Service, which can be downloaded from the website. The input form of the web interface (Figure 2a) is supported by Ajax to facilitate the identification of the protein accession number and the location of a mutation on the protein sequence or on the schematic 3D map provided. Given the input data, the MSV3d pipeline generates a multi-level characterization of the variant to be predicted. If a 3D model is available, these values are translated into prolog facts, which then become the input for the prediction service. Thanks to the Prolog engine, the deductive reasoning process immediately derives a conclusion (deleterious or neutral nsSNP) with identified rules. Figure 2b shows the KD4v output for the substitution Gly138Phe in the human peroxisomal biogenesis factor 3, predicted to be deleterious. In the 3D model of this protein, which is involved in the Zellweger syndrome, this residue is buried and located in one of the central helices shaping the protein fold. Analyzing the rule associated with this deleterious prediction, it can be seen that, although this residue is not highly conserved (67% identity which corresponds to the rank2 in our conservation pattern classification), the gain in hydrophobic contact and the decrease in the overall stability might be responsible for the deleterious effect.
Figure 2.
(a) Screenshot of the input form of the prediction service. (b) Screenshot of the output page providing the prediction results as well as the multi-level characterizations of the mutation. The rules are described if the variant is ‘deleterious’. The annotated information related to the mutated position can be visualized in the MSV3d interface on the right.
Prediction evaluation
We compared the performance of our ILP-based prediction service with two widely used methods: SIFT and PolyPhen-2. The different measures of predictive performance are reported for two independent nsSNP validation sets (Tables 1 and 2). The accuracy (72.28% in Table 1, 75.57% in Table 2) and F-measure (78.61% in Table 1, 71.52% in Table 2) indicate that the KD4v prediction service based on ILP is comparable to SIFT and PolyPhen-2 (although PolyPhen-2 is more accurate on one of the validation sets) and thus represents a competitive alternative solution. Moreover, the KD4v provides ILP rules associated with deleterious predictions that are more interpretable than the previous prediction methods. These rules should help to improve the understanding of the relationships between physico-chemical and structural features and deleterious mutations.
Table 1.
Comparison of prediction methods based on the PolyPhen-2 validation set [658 disease-causing (OMIM phenotype) mutations and 298 neutral polymorphisms]
| TP | FP | FN | TN | Sensitivity | Specificity | Precision | Recall | Accuracy | F-measure | |
|---|---|---|---|---|---|---|---|---|---|---|
| SIFT | 398 | 38 | 260 | 260 | 0.6049 | 0.8725 | 0.9128 | 0.6049 | 0.6883 | 0.7276 |
| PolyPhen-2 | 576 | 111 | 77 | 184 | 0.8821 | 0.6237 | 0.8384 | 0.8821 | 0.8017 | 0.8597 |
| KD4v | 487 | 94 | 171 | 204 | 0.7401 | 0.6846 | 0.8382 | 0.7401 | 0.7228 | 0.7861 |
Table 2.
Comparison of prediction methods based on the validation set that excludes proteins present in the training set (173 disease-causing mutations (OMIM phenotype) and 179 neutral polymorphisms)
| TP | FP | FN | TN | Sensitivity | Specificity | Precision | Recall | Accuracy | F-measure | |
|---|---|---|---|---|---|---|---|---|---|---|
| SIFT | 106 | 23 | 67 | 156 | 0.6127 | 0.8715 | 0.8217 | 0.6127 | 0.7443 | 0.702 |
| PolyPhen-2 | 139 | 70 | 34 | 109 | 0.8035 | 0.6089 | 0.6651 | 0.8035 | 0.7045 | 0.7278 |
| KD4v | 108 | 21 | 65 | 158 | 0.6243 | 0.8827 | 0.8372 | 0.6243 | 0.7557 | 0.7152 |
CONCLUSION
The KD4v server uses the available or modelled 3D structures and information provided by the MSV3d pipeline to characterize and predict the phenotypic effect of a mutation. The main advantages of KD4v are (i) valuable predicates and ILP rules associated with the predictions, allowing biologists to identify deleterious mutations and interpret the results, (ii) an ergonomic web interface, incorporating the comprehensive annotation of missense variants, complemented with a SOAP-based remote API for multiple predictions. Furthermore, the effects of any unknown missense variant (1 of approximately 32 000 000 variants corresponding to all positions of mapped 3D structures and all possible amino acid replacements) can be predicted upon request by the user. In the future, we will extend the background knowledge, first by adding structural surface topology descriptions (20) of the proteins, allowing the precise mapping of different functional regions such as the protein core and the non-interacting or interacting surfaces, and second, by integrating useful knowledge about the functional impact of missense variants from the SNPdbe database (21). Finally, we intend to enhance the prediction performance by combining ILP with other machine learning methods.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1 and 2, Supplementary Figure 1.
FUNDING
The work was performed within the framework of the Decrypthon program, co-funded by Association Française contre les Myopathies [AFM, 14390-15392]; IBM and Centre National de la Recherche Scientifique (CNRS); ANR [EvolHHuPro: BLAN07-1-198915 and Puzzle-Fit: 09-PIRI-0018-02]; Institute funds from the CNRS, INSERM, the Université de Strasbourg and the Vietnam Ministry of Education and Training (CT 322). Funding for Open access charge: ANR-10-BINF-03-02.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The IGBMC common services and BIPS platforms are acknowledged for their assistance.
REFERENCES
- 1.Thusberg J, Olatubosun A, Vihinen M. Performance of mutation pathogenicity prediction methods on missense variants. Hum. Mutat. 2011;32:358–368. doi: 10.1002/humu.21445. [DOI] [PubMed] [Google Scholar]
- 2.Ng PC, Henikoff S. SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thomas PD, Campbell MJ, Kejariwal A, Mi H, Karlak B, Daverman R, Diemer K, Muruganujan A, Narechania A. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 2003;13:2129–2141. doi: 10.1101/gr.772403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bromberg Y, Yachdav G, Rost B. SNAP predicts effect of mutations on protein function. Bioinformatics. 2008;24:2397–2398. doi: 10.1093/bioinformatics/btn435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Calabrese R, Capriotti E, Fariselli P, Martelli PL, Casadio R. Functional annotations improve the predictive score of human disease-related mutations in proteins. Hum. Mutat. 2009;30:1237–1244. doi: 10.1002/humu.21047. [DOI] [PubMed] [Google Scholar]
- 6.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bao L, Zhou M, Cui Y. nsSNPAnalyzer: identifying disease-associated nonsynonymous single nucleotide polymorphisms. Nucleic Acids Res. 2005;33:W480–W482. doi: 10.1093/nar/gki372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yue P, Melamud E, Moult J. SNPs3D: candidate gene and SNP selection for association studies. BMC Bioinformatics. 2006;7:166. doi: 10.1186/1471-2105-7-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Muggleton S. Inductive logic programming. N. Gen Comput. 1991;8:295–318. [Google Scholar]
- 10.Luu TD, Rusu AM, Walter V, Ripp R, Moulinier L, Muller J, Toursel T, Thompson JD, Poch O, Nguyen H. MSV3d: database of human MisSense variants mapped to 3D protein structure. Database J. Biol. Databases Curation. 2012;2012:bas018. doi: 10.1093/database/bas018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Friedrich A, Garnier N, Gagniere N, Nguyen H, Albou LP, Biancalana V, Bettler E, Deleage G, Lecompte O, Muller J, et al. SM2PH-db: an interactive system for the integrated analysis of phenotypic consequences of missense mutations in proteins involved in human genetic diseases. Hum. Mutat. 2010;31:127–135. doi: 10.1002/humu.21155. [DOI] [PubMed] [Google Scholar]
- 12.Plewniak F, Bianchetti L, Brelivet Y, Carles A, Chalmel F, Lecompte O, Mochel T, Moulinier L, Muller A, Muller J, et al. PipeAlign: A new toolkit for protein family analysis. Nucleic Acids Res. 2003;31:3829–3832. doi: 10.1093/nar/gkg518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Andreeva A, Howorth D, Chandonia JM, Brenner SE, Hubbard TJ, Chothia C, Murzin AG. Data growth and its impact on the SCOP database: new developments. Nucleic Acids Res. 2008;36:D419–D425. doi: 10.1093/nar/gkm993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thompson JD, Muller A, Waterhouse A, Procter J, Barton GJ, Plewniak F, Poch O. MACSIMS: multiple alignment of complete sequences information management system. BMC Bioinformatics. 2006;7:318. doi: 10.1186/1471-2105-7-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eswar N, Eramian D, Webb B, Shen MY, Sali A. Protein structure modeling with MODELLER. Methods Mol. Biol. 2008;426:145–159. doi: 10.1007/978-1-60327-058-8_8. [DOI] [PubMed] [Google Scholar]
- 16.Sobolev V, Sorokine A, Prilusky J, Abola EE, Edelman M. Automated analysis of interatomic contacts in proteins. Bioinformatics. 1999;15:327–332. doi: 10.1093/bioinformatics/15.4.327. [DOI] [PubMed] [Google Scholar]
- 17.Capriotti E, Fariselli P, Casadio R. I-Mutant2.0: predicting stability changes upon mutation from the protein sequence or structure. Nucleic Acids Res. 2005;33:W306–W310. doi: 10.1093/nar/gki375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mottaz A, David FP, Veuthey AL, Yip YL. Easy retrieval of single amino-acid polymorphisms and phenotype information using SwissVar. Bioinformatics. 2010;26:851–852. doi: 10.1093/bioinformatics/btq028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bard N, Bolze R, Caron E, Desprez F, Heymann M, Friedrich A, Moulinier L, Nguyen NH, Poch O, Toursel T. Decrypthon grid - grid resources dedicated to neuromuscular disorders. Stud. Health Technol. Informat. 2010;159:124–133. [PubMed] [Google Scholar]
- 20.Albou LP, Poch O, Moras D. M-ORBIS: mapping of molecular binding sites and surfaces. Nucleic Acids Res. 2011;39:30–43. doi: 10.1093/nar/gkq736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schaefer C, Meier A, Rost B, Bromberg Y. SNPdbe: constructing an nsSNP functional impacts database. Bioinformatics. 2012;28:601–602. doi: 10.1093/bioinformatics/btr705. [DOI] [PMC free article] [PubMed] [Google Scholar]