Abstract
Recent rapid advances in next generation RNA sequencing (RNA-Seq)-based provide researchers with unprecedentedly large data sets and open new perspectives in transcriptomics. Furthermore, RNA-Seq-based transcript profiling can be applied to non-model and newly discovered organisms because it does not require a predefined measuring platform (like e.g. microarrays). However, these novel technologies pose new challenges: the raw data need to be rigorously quality checked and filtered prior to analysis, and proper statistical methods have to be applied to extract biologically relevant information. Given the sheer volume of data, this is no trivial task and requires a combination of considerable technical resources along with bioinformatics expertise. To aid the individual researcher, we have developed RobiNA as an integrated solution that consolidates all steps of RNA-Seq-based differential gene-expression analysis in one user-friendly cross-platform application featuring a rich graphical user interface. RobiNA accepts raw FastQ files, SAM/BAM alignment files and counts tables as input. It supports quality checking, flexible filtering and statistical analysis of differential gene expression based on state-of-the art biostatistical methods developed in the R/Bioconductor projects. In-line help and a step-by-step manual guide users through the analysis. Installer packages for Mac OS X, Windows and Linux are available under the LGPL licence from http://mapman.gabipd.org/web/guest/robin.
INTRODUCTION
Next-generation high-throughput sequencing (NGS) is leading to the accumulation of a wealth of genomic and high-throughput mRNA sequencing (RNA-Seq) data is enabling increasingly comprehensive transcriptomic studies. A vast volume of expression data is being made available to the research community via several public data repositories, e.g. SRA (1) and ENA, (2). These advances have also greatly expanded the range of species amenable to transcriptomic analysis, by essentially providing a means to create new transcriptomes from the data itself. As described in several recent studies (3–6), long Roche/454 and short Illumina/Solexa or SOLiD sequencing reads can be used to first assemble a reference transcriptome of a hitherto poorly sequenced species and subsequently assess differential gene expression (DGE). Continual refinement of technologies and decreasing per-base sequencing costs will allow de novo sequencing approaches to be adopted by an increasing number of labs. These advances have created a demand for user-friendly software that enables researchers to handle NGS data sets and extract biologically relevant information.
Next-generation sequencing-based analysis of DGE is a multi-step process that includes raw data quality checking and filtering out of low quality data and contaminant sequences, mapping of the pre-processed reads to a reference, and statistical analysis of DGE to identify significantly responding genes. Several software tools have been developed to perform single steps in this workflow: FastQC is an excellent tool for generating quality overviews (http://www.bioinformatics.bbsrc.ac.uk/projects/fastqc), while FASTX provides a collection of command line programs to process and filter raw sequence data (http://hannonlab.cshl.edu/fastx_toolkit/). A range of specialized non-commercial aligners are available to map short sequence reads to a large reference genome or transcriptome [see (7) and (8) for a recent overview on algorithms and tools]. Finally, methods for statistical inference of DGE from mapped RNA-Seq reads have been established in the Bioconductor project (9) are being continuously developed by leading biostatisticians, including edgeR (10), DESeq (11) and baySeq (12). These packages assume a negative binomial distribution of the RNA-Seq count data but use slightly different approaches for the inference of DGE, providing an excellent framework for RNA-Seq-based transcript profiling.
Although tools are available to perform individual steps in RNA-Seq analysis, it is not trivial to use them for a complete pipeline. Many of the programs only provide command line interfaces. They are sometimes not directly compatible with respect to their input/output file formats. Hence, running a complete RNA-Seq-based DGE analysis requires considerable bioinformatics skills. This is an obstacle for many non-specialist researchers.
To date, few non-commercial applications featuring a graphical user interface (GUI) are available for RNA-Seq analysis. Most of these are not distributed as stand-alone tools and require a complicated installation and setup. GenePattern (13), for example, provides a very versatile collection of analysis functions including DGE, SNP and proteomics analyses. Tools like Myrna (14) and Galaxy (15–17) take advantage of cloud and cluster computing to boost performance when processing large data volumes, but rely on an elaborate bioinformatic infrastructure and lack an intuitive user interface. They constitute excellent and flexible analysis platforms for use in bioinformatics units but are less suitable for non-specialists. SAMMate (18) is a stand-alone graphical workbench-like application providing NGS analysis functions that are also needed for RNA-Seq analysis. However its GUI does not follow a workflow-oriented step-by-step process.
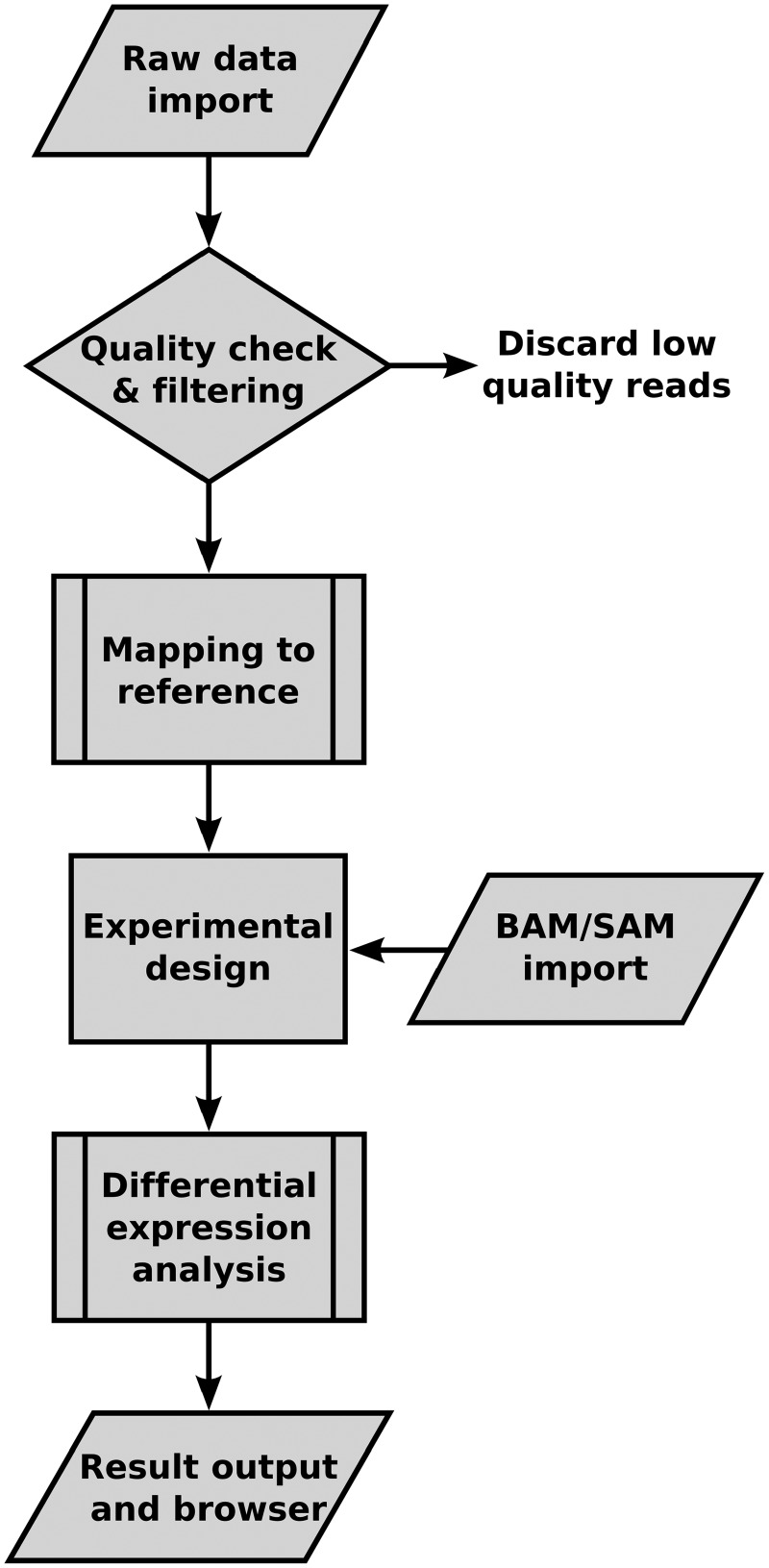
We have developed RobiNA as an integrated, cross-platform application that provides user-friendly workflows and guides the user through each step of DGE analysis (see Figure 1 for an overview). RobiNA allows users to import short read data in FastQ format and do thorough quality assessment and filtering prior to mapping the reads to a user-provided reference genome or transcriptome. The mapping of reads is based on the open source BOWTIE alignment tool (19), which is fully integrated in the RobiNA application package. The last step is statistical analysis of DGE based on the Bioconductor packages edgeR and DESeq. The R statistics software engine and all required Bioconductor packages are integrated into the RobiNA application package, making installation and configuration of external tools unnecessary on the commonly used operating systems, Windows and Mac OS X. On Linux, RobiNA requires a working installation of R version 2.15.0 or higher. RobiNA is distributed under the LGPL licence as all-in-one installer packages that contain all necessary software tools plus a manual explaining the analysis workflows step-by-step. The packages, manual and demo datasets are available for download from http://mapman.gabipd.org/web/guest/robin.
Figure 1.
Flow chart of RNA-Seq-based differential gene-expression analysis steps provided by RobiNA.
RNA-SEQ WORKFLOW
Data import
Raw Illumina/Solexa short read data can be imported in FastQ format. RobiNA will determine the quality encoding version of the input data based on a sample taken from each input file. This is necessary to make sure that differences in the way the quality scores were generated in the Illumina data processing pipeline are properly taken into account during checking and filtering. Alternatively, users can import BAM/SAM alignment files. If this option is chosen, the quality checking and filtering steps can be omitted and the workflow is shortened to the experiment design and statistical analysis steps. While input files can be bzip2- or gzip-compressed, we recommend using uncompressed files as some quality checking options are not available for compressed data, and the data need to be uncompressed prior to the mapping step anyway. Users wishing to reanalyse already pre-computed counts tables [as publicly available from e.g. ReCount repository (20)] can import these as plain text files by activating the alternative import option and directly move forward to the statistical analysis step.
Quality checking and read filtering
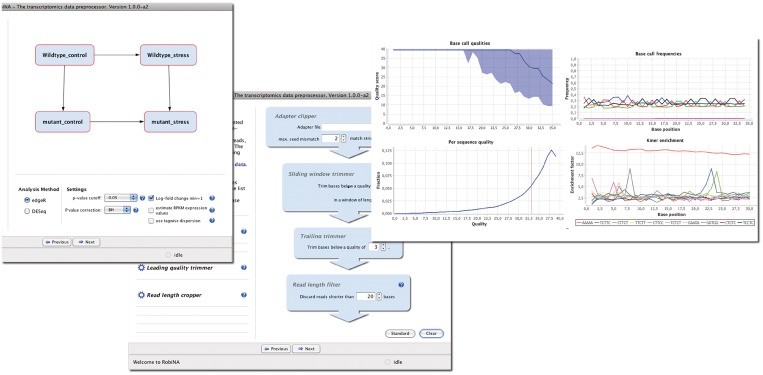
RobinNA provides a range of quality checking modules covering different aspects of raw read quality. These can be freely combined to gain a broad overview of the input data (Figure 2). The selected modules are applied to each input file separately, allowing the user to identify and potentially exclude low quality sequencing runs. Specifically, RobiNA provides the following six quality assessment modules: (i) Base call quality summary. The base call quality scores that are assigned to each nucleotide during the base calling step of the NGS pipeline are summarized in plots showing the median and the 25th and 75th percentile score at each nucleotide position across all reads. Positions at which the quality drops below a score of 13 (i.e. error probability of P ∼ 0.05) are highlighted in red. The global base call quality distribution, computed based on the mean quality score of each read, is shown in a second plot that also gives the overall mean quality score. (ii) Base call frequencies. Nucleotide base frequencies are computed across all reads at each position and shown as a combined line graph. Ideally, these curves should be almost level and smooth lines, which mirror, at each position, the overall base composition of the examined organism. Peaks of individual nucleotides at a given position indicate a substantial bias, and are often observed when barcode or adapter sequences have not been fully excluded. (iii) Consecutive homopolymers. In a rarely observed but serious technical artefact, which we term ‘consecutive homopolymer error’, all the bases in a window of several bases starting from the same position in each read are identical to the preceding base. This artefact shows up as a peak in the homopolymer fraction at the corresponding positions and is visualized in a line plot. (iv) K-mers. This module scans the reads for short sequences of k nucleotides (K-mers) that occur more often than expected based on the nucleotide composition of the analyzed reads. By default, RobiNA scans for 5-mers and records up to 106 unique K-mers. These settings were chosen to keep memory usage low. Users have the option to scan a range from k = 5 to k = 10. K-mers observed three times more often than expected by chance are reported in a table, and their positional enrichment across all analyzed reads is shown in a multiple line graph. Overenriched K-mers are very often indicative of contamination of the sequence with adapters or barcodes. Low quality sequence data will also frequently exhibit an overenrichment of homopolymer K-mers towards the end of the reads. (v) Overenriched sequences. Similar to the K-mer frequency analysis, RobiNA screens for frequently occurring longer sequence stretches. These are usually due to adapter sequences used in the sequencing library preparation process and should be removed in the subsequent filtering step. (vi) Basic statistics provides a general overview of the data by computing base statistics such as the global nucleotide composition, number of reads and bases and the number of failed base calls (‘N’ content).
Figure 2.
Screen shots showing excerpts of RobiNAs GUI. The left panel shows the experiment designer step that allows the graphical definition of comparisons of interest. The middle panel illustrates the trimming pipeline setup. On the right side examples of quality check plots are shown. The panel shows a base call quality summary plot (upper left), positional base call frequencies (upper right), overall read quality distribution (lower left) and the positional K-mer enrichment plot (lower right).
Quality checking can take a substantial amount of time on slower computers. When tested on an iMac with a 2.4 GHz Intel Core 2 duo CPU and 2 GB of RAM (running Mac OS X 10.6.8), quality checking of a 1 GB FastQ file containing 35-nucl long Illumina/Solexa reads took approximately 5 min. Trimming of the same file took about 10 min with the adapter clipper, sliding window trimmer and length filter modules activated. RobiNA accelerates the process by running several quality checks in parallel in separate threads. The number of parallel processes is initially set to the number of CPUs detected on the computer but can be modified by the user. Additionally, when using uncompressed input data, users can save time by running quality checks on a random sample of the input data. The sample size can be modified in the ‘File settings’ tab in the quality check settings step. Depending on the sample size this will give a fast (but less representative if small sample sizes are used) overview of the input data quality. The quality check results can be immediately browsed in RobiNA. They are automatically saved to the analysis project folder as PDF files when proceeding to the filtering step.
The filtering step is organized as a modular construction kit. Seven different filter modules can be freely combined to build a custom read trimming and filtering pipeline. A range of modules is provided: quality-based trimmers remove low-quality bases from the start and end of each read or by scanning across each read with a sliding window. A read length cropper can be used to shorten all reads to a specified length. Reads that are too short are removed by a minimal length filter. Known adapter sequences supplied by the user can be removed using the adapter clipper module. A barcode splitter module divides multiplexed barcoded reads into separate files. A custom trimming pipeline can be assembled simply by drag & dropping modules into a workflow area. Each trimming step is represented by a small GUI that displays all modifiable trimming parameters to the user (Figure 2). We also make the trimming pipeline available as a stand-alone command line tool called Trimmomatic. The detailed description of this module will, due to space constraints, be presented in a separate publication (Bolger et al.., manuscript in preparation).
Read library setup and reference mapping
In the next step, the reads are mapped to a reference sequence. This must be supplied by the user as a FASTA file of all transcripts or a FASTA file plus matching GFF3 annotation, depending on whether the reference is transcriptomic or genomic reference data.
The mapping step requires prior definition of the layout of the experiment in the ‘Experiment layout’ step. A visual interface allows the user to enter the different treatments and define which trimmed read file represents a sample of which treatment. Even though the downstream statistical analysis supports experiments with only one replicate per treatment, it is strongly recommended to provide more than one biological replicate to make sure that the results are reliable.
After this, the mapping process starts. Filtered reads are aligned to the reference using the BOWTIE aligner. BOWTIE is included in the application packages. When a new reference sequence is used for the first time, a BOWTIE search index is built and saved for subsequent usage. Basic quality statistics of the reference sequence such as N50, N content, number of sequences and genes and the average sequence length are recorded.
The accuracy can be influenced by modifying the settings of the BOWTIE aligner. RobiNA provides two preset configurations that, allow different degrees of mismatch in the alignments. By default, no mismatch is tolerated in a seed region of 28 nt at the beginning of the reads. However, a more permissive setting might be justified in some circumstances, for example when working with reads originating from a strain that differs from the reference strain. By choosing the ‘custom’ setting, users can freely modify the number of allowed mismatches, the length of the alignment seed region and the tolerated sum of mismatch quality scores to adapt the mapping process to their specific needs. However, only unique alignments will be recorded and used for counting gene abundances for DGE analysis.
RobiNA offers the option to compute normalized estimates of the expression level of each gene expressed as estimated RPKM values (reads per kilobase of exon model per million mapped reads). RPKM values are computed based on the uniquely aligning reads only. In cases where a read maps into a genomic region where two genes overlap (e.g. genes on opposite strands), the shared reads are split between the genes weighted according to each gene’s expression level computed from unambiguously mapping reads. The RPKM values, however, are provided as rough estimates of gene expression only and are not used in the DGE analysis.
Experiment designer and statistical analysis
Gene abundances are recorded in a counts table listing the number of reads unambiguously mapped to each gene or transcript. At this stage, the user has to further formulate his experimental question by defining which treatments are to be compared with each other. This is done on the experiment designer panel, which is displayed when the mapping step has been completed. Each group of biological replicates of a treatment is represented by a blue box. Users can define any number of (non-redundant) direct comparisons of treatments by connecting two boxes with an arrow by clicking on one box and then holding down the control key and dragging the mouse to the other box (Figure 2). As soon as the mouse button is released, the comparison is defined as ‘Treatment A minus Treatment B’. Genes that show a higher or lower expression in treatment A will have a positive and negative log fold-change, respectively.
Statistical inference of DGE is initiated by clicking ‘next’. The user can choose which method is used for the statistical DGE analysis and modify parameters that are relevant for the analysis. There is a choice of methods to correct computed raw P-values for multiple testing. The user can define cut-off P-values and choose to ignore genes with a log2-fold change <1 in the analysis.
Since it has been shown that the GC content can have a substantial impact on the read abundances in a RNA-Seq data set (21–23) we incorporated the GC content bias cor- rection methods implemented in the EDASeq package (24). After activating the GC content correction option, users can choose which method to use for within-lane and between-lane normalization. If no GC bias correction is performed, RobiNA will perform the default normalization steps of the selected differential expression analysis package.
RobiNA uses the excellent edgeR and DESeq packages developed in the Bioconductor project. All user input and the counts table generated in the mapping step is used to generate an R script that executes the statistical analysis. The script is saved, together with all other results, in the project folder and can be inspected and re-run independently. The output of the statistical analysis is a set of detailed tables giving log fold changes and P-values for differential expression for each comparison, a condensed results file that combines the results of all comparisons in one table, and a range of descriptive plots that provide an overview of the results. Additionally, all quality checking results, intermediary mapping results (lists of unique and ambiguous reads for each sample) and log files documenting the trimming and overall workflow progress plus a PDF summary file are saved in the project folder. RobiNA-generated scripts can serve as a convenient starting point for further customized analyses by users who are experienced in the use of R/Bioconductor.
When working with plant data, users can choose to functionally annotate the data based on MapMan BINs (25) in a last step. A choice of MapMan functional annotation packages is provided in the RobiNA package. More mapping files can be freely downloaded from http://mapman.gabipd.org/web/guest/mapmanstore.
IMPLEMENTATION
RobiNA is implemented in Java and R and contains an R engine plus all R packages required to run the statistical analyses. BOWTIE binaries for Mac OS X, Windows and Linux have been added to the application package and are used for the mapping of short reads to reference sequences. In addition to the RNA-Seq-based analysis, RobiNA provides workflows for microarray analysis based on the previously published Robin tool (26). RobiNA makes use of several open source Java libraries. Specifically, the NetBeans visual API (http://graph.netbeans.org/) was used to develop the visual experiment designer, and Apache commons (http://commons.apache.org/) was used to facilitate generic string operations. To achieve an improved user experience and better integration into the Mac OS X platform, we used the AppleJavaExtensions provided by Apple, Inc., and the ‘QuaQua look and feel’ (http://www.randelshofer.ch/quaqua/). The SAM JDK library (http://picard.sourceforge.net/) is used for import of SAM/BAM files, and libraries developed by the biojava project (27) are used for working with GFF3 annotation files. Bzip2 support is provided by http://code.google.com/p/jbzip2/. Generation of plots is based on JFreeChart (http://www.jfree.org/jfreechart/) and PDF output is provided by iTextPDF (http://itextpdf.com/).
Installer packages for different operating systems were created using the free IzPack installer generator (http://izpack.org/). We also provide a lightweight package without R that can be deployed on any Java-enabled platform. On first use, this version of RobiNA will ask the user for a path to a working R installation, check this installation and automatically download all required packages (if not already present), provided the computer has a working internet connection.
CONCLUSIONS
Next generation RNA sequencing greatly extends the possibilities of transcript profiling. We have developed RobiNA as a user-friendly all-in-one application that enables researchers to perform all steps of the analysis in a flexible yet user friendly way. To our knowledge, RobiNA is the first application providing a complete stand-alone RNA-Seq-based DGE analysis workflow. We believe it will be a useful tool for the community to cope with this new technology.
FUNDING
Funding for open access charge: The Max Planck Society; RobiNA was developed within the Plant KBBE-SAFQIM, the German Ministry of Education and Research (BMBF) [project 0315912].
Conflict of interest statement. None declared.
REFERENCES
- 1.Wheeler DL, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, Dicuccio M, Edgar R, Federhen S, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2008;36:D13–D21. doi: 10.1093/nar/gkm1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leinonen R, Akhtar R, Birney E, Bower L, Cerdeno-Tarraga A, Cheng Y, Cleland I, Faruque N, Goodgame N, Gibson R, et al. The European Nucleotide Archive. Nucleic Acids Res. 2010;39:D28–D31. doi: 10.1093/nar/gkq967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bajgain P, Richardson BA, Price JC, Cronn RC, Udall JA. Transcriptome characterization and polymorphism detection between subspecies of big sagebrush (Artemisia tridentata) BMC Genomics. 2011;12:370. doi: 10.1186/1471-2164-12-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang TY, Chen HL, Lu MY, Chen YC, Sung HM, Mao CT, Cho HY, Ke HM, Hwa TY, Ruan SK, et al. Functional characterization of cellulases identified from the cow rumen fungus neocallimastix patriciarum W5 by transcriptomic and secretomic analyses. Biotechnol. Biofuels. 2011;4:24. doi: 10.1186/1754-6834-4-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Siebert S, Robinson MD, Tintori SC, Goetz F, Helm RR, Smith SA, Shaner N, Haddock SH, Dunn CW. Differential gene expression in the Siphonophore Nanomia bijuga (Cnidaria) assessed with multiple next-generation sequencing workflows. PloS One. 2011;6:e22953. doi: 10.1371/journal.pone.0022953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Su CL, Chao YT, Alex Chang YC, Chen WC, Chen CY, Lee AY, Hwa KT, Shih MC. De novo assembly of expressed transcripts and global analysis of phalaenopsis aphrodite transcriptome. Plant Cell Physiol. 2011;52:1501–1514. doi: 10.1093/pcp/pcr097. [DOI] [PubMed] [Google Scholar]
- 7.Li H, Homer N. A survey of sequence alignment algorithms for next-generation sequencing. Brief Bioinform. 2010;11:473–483. doi: 10.1093/bib/bbq015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oshlack A, Robinson MD, Young MD. From RNA-seq reads to differential expression results. Genome Biol. 2010;11:220. doi: 10.1186/gb-2010-11-12-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2009;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hardcastle TJ, Kelly KA. baySeq: empirical Bayesian methods for identifying differential expression in sequence count data. BMC Bioinformatics. 2010;11:422. doi: 10.1186/1471-2105-11-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reich M, Liefeld T, Gould J, Lerner J, Tamayo P, Mesirov JP. GenePattern 2.0. Nat Genet. 2006;38:500–501. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- 14.Langmead B, Hansen KD, Leek JT. Cloud-scale RNA-sequencing differential expression analysis with Myrna. Genome Biol. 2010;11:R83. doi: 10.1186/gb-2010-11-8-r83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goecks J, Nekrutenko A, Taylor J. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 2010;11:R86. doi: 10.1186/gb-2010-11-8-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giardine B, Riemer C, Hardison RC, Burhans R, Elnitski L, Shah P, Zhang Y, Blankenberg D, Albert I, Taylor J, et al. Galaxy: a platform for interactive large-scale genome analysis. Genome Res. 2005;15:1451–1455. doi: 10.1101/gr.4086505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blankenberg D, Von Kuster G, Coraor N, Ananda G, Lazarus R, Mangan M, Nekrutenko A, Taylor J. Galaxy: a web-based genome analysis tool for experimentalists. Curr Protoc Mol Biol. 2010;89:19.10.1–19.10.21. doi: 10.1002/0471142727.mb1910s89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu G, Deng N, Zhao Z, Judeh T, Flemington E, Zhu D. SAMMate: a GUI tool for processing short read alignments in SAM/BAM format. Source Code Biol Med. 2011;6:2. doi: 10.1186/1751-0473-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frazee AC, Langmead B, Leek JT. ReCount: a multi-experiment resource of analysis-ready RNA-seq gene count datasets. BMC Bioinformatics. 2011;12:449. doi: 10.1186/1471-2105-12-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hansen KD, Brenner SE, Dudoit S. Biases in Illumina transcriptome sequencing caused by random hexamer priming. Nucleic Acids Res. 2010;38:e131. doi: 10.1093/nar/gkq224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bullard JH, Purdom E, Hansen KD, Dudoit S. Evaluation of statistical methods for normalization and differential expression in mRNA-Seq experiments. BMC Bioinformatics. 2010;18:94. doi: 10.1186/1471-2105-11-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Benjamini Y, Speed TP. Summarizing and correcting the GC content bias in high-throughput sequencing. Nucleic Acids Res. 2012;40:e72. doi: 10.1093/nar/gks001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Risso D, Schwartz K, Sherlock G, Dudoit S. GC-content normalization for RNA-Seq data. BMC Bioinformatics. 2011;12:480. doi: 10.1186/1471-2105-12-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Usadel B, Poree F, Nagel A, Lohse M, Czedik-Eysenberg A, Stitt M. A guide to using MapMan to visualize and compare Omics data in plants: a case study in the crop species, Maize. Plant Cell Environ. 2009;32:1211–1229. doi: 10.1111/j.1365-3040.2009.01978.x. [DOI] [PubMed] [Google Scholar]
- 26.Lohse M, Nunes-Nesi A, Kruger P, Nagel A, Hannemann J, Giorgi FM, Childs L, Osorio S, Walther D, Selbig J, et al. Robin: an intuitive wizard application for R-based expression microarray quality assessment and analysis. Plant Physiol. 2010;153:642–651. doi: 10.1104/pp.109.152553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Holland RC, Down TA, Pocock M, Prlic A, Huen D, James K, Foisy S, Drager A, Yates A, Heuer M, et al. BioJava: an open-source framework for bioinformatics. Bioinformatics. 2008;24:2096–2097. doi: 10.1093/bioinformatics/btn397. [DOI] [PMC free article] [PubMed] [Google Scholar]