Abstract
The Sorting Intolerant from Tolerant (SIFT) algorithm predicts the effect of coding variants on protein function. It was first introduced in 2001, with a corresponding website that provides users with predictions on their variants. Since its release, SIFT has become one of the standard tools for characterizing missense variation. We have updated SIFT’s genome-wide prediction tool since our last publication in 2009, and added new features to the insertion/deletion (indel) tool. We also show accuracy metrics on independent data sets. The original developers have hosted the SIFT web server at FHCRC, JCVI and the web server is currently located at BII. The URL is http://sift-dna.org (24 May 2012, date last accessed).
INTRODUCTION
An individual’s genome contains approximately 3.7 million single nucleotide variants (SNVs) which can be identified by whole-genome sequencing (1). The challenge for geneticists is to identify what are the causal variants for the phenotype or disease being studied. The majority of SNVs found in a human are common among the population, but disease-causing variants are typically private or rare, and tend to occur in protein coding regions which constitute only 1% (30 megabases) of the total genome (2,3). Databases like dbSNP (4) and 1000 Genomes (5) are useful for filtering out common variants, but the remaining variants need to be sorted and prioritized to identify those that may potentially affect protein function. Algorithms like SIFT can help in this respect.
Sorting Intolerant from Tolerant (SIFT) is an algorithm that predicts the potential impact of amino acid substitutions on protein function. We have recently extended SIFT to predict on frameshifting indels (6). For amino acid substitutions, SIFT has been used actively in human genetic research (7–9) [e.g. cancer (10,11) Mendelian diseases (12) and infectious diseases (13)]. We emphasize that SIFT’s utility extends beyond research on humans and human disease studies. SIFT has been used to study the effects of missense mutations on agricultural plants (14,15), and model organisms like rats (16,17), canines (18) and Arabidopsis (19). In general, SIFT is useful in cases where research work involves filtering through a plethora of SNVs and indels to identify causal variants.
MATERIALS AND METHODS
The human variation (HumVar) and human divergence (HumDiv) data sets used to assess SIFT’s performance were obtained from UniProtKB (20). Adzhubei et al. (20) compiled the HumDiv deleterious list using mutations annotated to cause Mendelian diseases in humans. They created the HumDiv neutral data set by comparing human proteins to their homologs in closely related mammals, and identifying amino acids that are different. For the HumVar deleterious data set, the authors included any mutation annotated to cause human disease, regardless of whether they are Mendelian in origin or not. The HumVar neutral data set is made up of nonsynonymous polymorphisms not annotated as disease causing. We mapped the HumVar and HumDiv data to Ensembl, RefSeq and UCSC Known ids using the UniProtKB id mapping tool (http://www.uniprot.org/help/uniprotkb). Not all mutations from the data sets could be mapped. Hence, the final number of mutations used is less than that of the original dataset (Table 1). True positives (TP) are defined as disease-causing mutations correctly predicted to affect protein function, and false negatives (FN) are those incorrectly predicted to be tolerated. True negatives (TN) are neutral variations correctly predicted as tolerated and false positives (FP) are neutral variations incorrectly predicted to affect protein function.
Table 1.
Number of HumDiv and HumVar data points used to assess SIFT’s performance
| Data set | Number of data points |
Coverage** (%) | ||
|---|---|---|---|---|
| From original dataset (20) | Used in evaluating SIFT* | With SIFT predictions | ||
| HumDiv neutral | 6027 | 5816 | 5582 | 96.0 |
| HumDiv deleterious | 3055 | 2893 | 2791 | 96.5 |
| HumVar neutral | 8638 | 7475 | 7178 | 96.0 |
| HumVar deleterious | 12 598 | 11 982 | 11 561 | 96.5 |
*Lookups to the SIFT database required Ensembl, RefSeq and UCSC Known protein identifiers and the chromosome associated with the given identifier. Not all data points could be mapped to these types of protein identifiers using UniProtKB’s ID mapping tool. Furthermore, we were not able to map some proteins to their chromosomes.
**Coverage = (Number with predictions/Number of data points tested)
The various statistics are computed as follows:
Sensitivity = TP/(TP + FN)
Specificity = TN/(TN + FP)
Accuracy = (TP + TN) / (TP + TN + FP + FN)
Precision = TP / (TP + FP)
Negative predictive value (NPV) = TN / (TN + FN)
Matthews correlation coefficient (MCC) = X / Y
where X = [(TP × TN) – (FP × FN)] and Y = SQRT[(TP + FP) (TP + FN) (TN + FP) (TN + FN)].
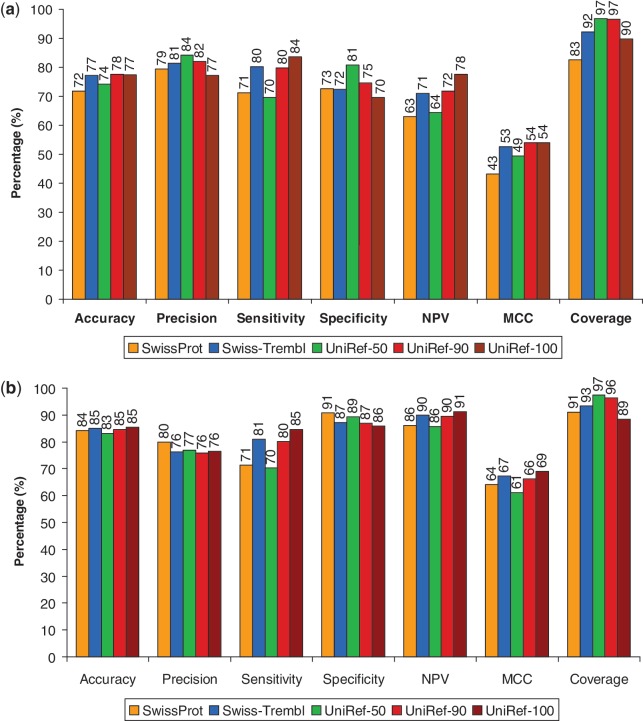
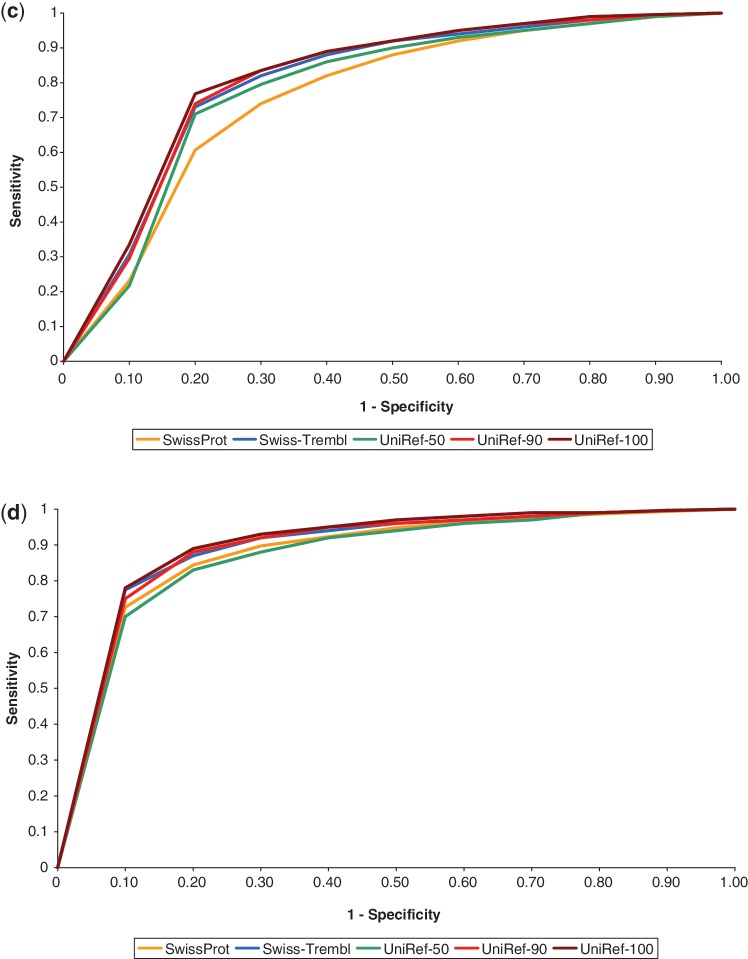
We generated receiver operating characteristic (ROC) curves for each protein database by computing the SIFT score for each substitution and categorizing them as tolerated or deleterious using different threshold values. For each threshold, the true positive rate (sensitivity) and false positive rate (1 – specificity) are then computed and plotted in Figure 1.
Figure 1.
Performance statistics of SIFT predictions on PolyPhen-2’s (a) HumVar and (b) HumDiv data sets when using various protein databases. ROC curves on the (c) HumVar and (d) HumDiv data sets. Although UniRef-100 shows slightly better performance than UniRef-90, it has lower coverage.
RESULTS
Algorithm description
SIFT uses sequence homology to compute the likelihood that an amino acid substitution will have an adverse effect on protein function. The underlying assumption is that evolutionarily conserved regions tend to be less tolerant of mutations, and hence amino acid substitutions or insertions/deletions in these regions are more likely to affect function.
The SIFT workflow begins with a query protein that is searched against a protein database to obtain homologous protein sequences. Sequences with appropriate sequence diversity are chosen (21). The chosen sequences are aligned, and for a particular position, SIFT looks at the composition of amino acids and computes the score. A SIFT score is a normalized probability of observing the new amino acid at that position, and ranges from 0 to 1. A value of between 0 and 0.05 is predicted to affect protein function. Details of the algorithm can be found in (21,22).
The field of predicting the functional effects of nonsynonymous variations is long established. PolyPhen (23) and SIFT are two of the earlier works in this field. A recent paper lists more than 40 programs that provide similar functionality (24). Some of these algorithms have been directly compared to each other (25). At least two methods, MutPred (26) and nsSNPAnalyzer (27) incorporate the SIFT algorithm into their prediction pipelines, while BSIFT (28) uses the SIFT algorithm to include predictions for activating mutations.
SIFT PREDICTION PERFORMANCE
In this section, we assess SIFT’s performance on external data sets independent of the initial training sets. SIFT was originally trained and tested on LacI, lysozyme and HIV protease substitutions (21,22). We assess its performance on the HumDiv and HumVar data sets, which were created by the authors of PolyPhen-2 (20). In addition, we evaluate the effects of using different protein databases on prediction accuracy. This was motivated by Hicks et al. (29) who reported that prediction accuracy depends on the number of sequences and the sequence alignment. As the number of sequences chosen by the SIFT algorithm depends on the protein database used, we tested five protein databases (Swiss-Prot, Swiss-Prot with Trembl, UniRef-50, UniRef-90 and UniRef-100) and measured the resulting prediction accuracies.
Prediction accuracies were similar among the databases, but sensitivity and specificity can vary depending on the protein database used (Figure 1). Based on these results, we chose to use UniRef90 for our pre-computed SIFT scores due to its high coverage, high sensitivity and balanced performance.
Hicks et al. noted that SIFT exhibited low specificity when tested on 267 amino acid changes located in four genes (29). We applied our chosen protein database on these genes and evaluated SIFT’s performance. Table 2 compares the sensitivity and specificity reported by Hicks et al. against our results. For the 267 variants tested, sensitivity remains high, while specificity increased slightly. Similar algorithms (PolyPhen-2, Xvar) also showed high sensitivity and low specificity in the Hicks et al. paper (29). Users may prefer high sensitivity over high specificity so as not to miss true deleterious mutations. Hicks et al. noted that the genes in the test set can affect the accuracy of prediction algorithms, and this may be a factor resulting in lower specificity for these particular four genes. SIFT exhibited higher specificity on the HumDiv and HumVar data sets (Figure 1). Performance for prediction algorithms may differ for different data sets.
Table 2.
Comparison of SIFT’s performance on our predictions based on UniRef90 and that reported by Hicks et al.
| SIFT sensitivity (%) |
SIFT specificity (%) |
|||
|---|---|---|---|---|
| As reported by Hicks et al. (29) (%) | Generated using UniRef90 (%) | As reported by Hicks et al. (29) (%) | Generated using UniRef90 (%) | |
| MLH1 (60) | 72 | 92 | 52 | 57 |
| MSH2 (30) | 89 | 89 | 46 | 36 |
| TP53 (144) | 84 | 79 | 75 | 100 |
| BRCA1 (33) | 94 | 88 | 31 | 44 |
| Overall | 83 | 83 | 46 | 52 |
In the first column, numbers in parenthesis refers to the number of amino acid substitutions. Hicks et al. did not report accuracy and precision statistics and these are therefore not compared.
NEW FEATURES OF THE WEB SERVER
SIFT was one of the first amino acid prediction tools to have a web server (22). We have incorporated various ways to submit inputs to the SIFT web server for predictions (e.g. protein ids, batch protein and protein sequence submissions) (30,31). In this section, we discuss new features that have not been described previously.
Genome-wide database of predictions for nonsynonymous variants
Human genome sequences are being generated at a rapid rate. After read mapping and variant calling, the last step in characterizing a human genome is to annotate possible functional variants. For each protein sequence, SIFT takes approximately 10–20 min to run. A human genome contains approximately 8000–10 000 non synonymous variants (2), which would take a long time to generate SIFT predictions if the entire procedure was executed each time. Therefore, we speed up the process by (i) mutating every coding base in the reference genome to the other three possible DNA bases, (ii) computing the SIFT score for the resulting amino acid changes and (iii) finally storing the amino acid changes and their respective predictions in a database. When a user submits a list of genome variants, a simple lookup returns the predictions. Our most recent database provides predictions for RefSeq, UCSC, CCDS and Ensembl gene annotations.
Our current database contains approximately 79 × 106 unique nonsynonymous variations, out of which nearly 76 × 106 have prediction scores. The remainder are either (i) nonsense mutations, (ii) short protein sequences which could not be processed by SIFT or (iii) positions at the start or end of the protein which may not have enough sequences after the SIFT alignment procedure for prediction.
Variant annotation
Variants can be annotated with dbSNP and/or 1000 Genomes. Users can opt to have their queries annotated by dbSNP id with allele frequencies from the HapMap Project (32). We have also added the option to display 1000 Genomes with population allele frequencies (5). We chose to display HapMap and 1000 Genomes allele frequencies because they are derived from known populations of ‘normal’ individuals. dbSNP is known to contain false positives (21), hence allele frequencies can help to verify true variants in the human population. dbSNP incorporates 1000 Genomes data, but their lag time motivated us to incorporate 1000 Genomes separately. We will phase out 1000 Genomes annotation when it has been fully incorporated into dbSNP.
File format conversion
The input format for genomic variants into SIFT was established prior to next-generation sequencing. Since then, there are standard formats released by the next-generation sequencing software, and we have added the ability to convert different file formats, including VCF, Pileup and GFF files to the SIFT format. The conversion tool also filters out coordinates in non-coding regions, and returns the subset of coding positions. This speeds up the subsequent lookup process.
Indel prediction tool
Small indels (insertions/deletions of 20 bp or less) are the second largest class of mutations that lead to Mendelian diseases (33). We provided indel annotation (e.g. whether the indel causes nonsense-mediated decay, the indel’s effect on protein sequence) using VariantClassifier (34). We have also developed SIFT Indel, a prediction method for frameshifting indels as an extension to the SIFT algorithm (6). SIFT Indel was trained on a set of disease-causing frameshifting indels (3) and neutral indels derived from the UCSC pairwise alignments of the human genome with mammalian genomes (35). The tool was based on a decision tree algorithm using four features describing each indel and its influences on the gene product. The algorithm achieved 84% accuracy, with 90% sensitivity and 81% precision using 10-fold cross-validation. We applied SIFT Indel to human frameshifting indels and found that the percentage of frameshifting indels predicted to be deleterious is negatively correlated with allele frequency. This is a similar trend that has been previously seen for nonsynonymous SNPs, but for indels the effect is more severe. The server takes around 10–15 min to make predictions for 1000 indels.
Cloud
The SIFT source code and Linux executables are publicly available. A public image of SIFT is also available on the Amazon cloud so that users can run pre-installed SIFT directly. A step-by-step guide can be found on the website.
COMPARISON TO JCVI WEB SERVER
The current web server supported by the original authors of SIFT can be found at http://sift-dna.org. J. Craig Venter Institute (JCVI), where the original author of SIFT previously worked, maintains a separate SIFT web server that is independent of the one described here. JCVI has released its own versions: SIFT 4.0.3b and JCVI-SIFT 1.0.2. Users who utilize either of the web servers should understand that the performance of the tools may differ as they are managed by different groups.
We assessed the SIFT 4.0.3b August 2011 database and our November 2011 database. Although scores are not identical, similar accuracies are observed (Supplemental Figure S1). This is expected because different protein databases (e.g. Swiss-Prot, UniRef) were used to compute SIFT scores. This creates minor differences in scores but does not have a discernible impact on prediction performance, as shown in Figure 1.
The two databases differ in coverage. In our most recent release, we calculated predictions for RefSeq, CCDS and UCSC Known genes in addition to Ensembl gene annotations. Therefore, we have 1.95 million more missense predictions than the JCVI August 2011 database which uses Ensembl only (Supplemental Figure S2). In addition, the JCVI database is currently missing scores for chromosome Y (Supplemental Figure S2). More comparison details are described at sift-dna.org.
DISCUSSION
The SIFT web server (http://sift-dna.org) offers tools to predict the effects of nonsynonymous single nucleotide variants (nsSNVs) and frameshifting indels on protein function. In addition, a standalone version of the software can be downloaded or accessed through Amazon cloud. SIFT is useful for researchers who are interested in investigating the effects of mutations on protein functions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–2.
FUNDING
SIFT was funded by National Institutes of Health [RO1 GM068488] when hosted at FHCRC and by National Human Genome Research Institute [R01 HG004701] when SIFT was hosted at JCVI. Since June 2010, A*STAR has provided funding on work done on SIFT at the Genome Institute of Singapore, and it is hosted at the Bioinformatics Institute of Singapore. Funding for open access charge: Genome Institute of Singapore (A*STAR).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
SIFT is freely available to the academic community; commercial licensees should contact Steven Henikoff. We thank Tai Pang Yong, Jorja Henikoff, Lakshmi Radhakrishnan, Anthony Youzhi Cheng, and Qiangze Hoi for assistance in maintaining the SIFT server.
REFERENCES
- 1.Lam HY, Clark MJ, Chen R, Chen R, Natsoulis G, O'Huallachain M, Dewey FE, Habegger L, Ashley EA, Gerstein MB, et al. Performance comparison of whole-genome sequencing platforms. Nat. Biotechnol. 2011;30:78–82. doi: 10.1038/nbt.2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ng SB, Turner EH, Robertson PD, Flygare SD, Bigham AW, Lee C, Shaffer T, Wong M, Bhattacharjee A, Eichler EE, et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature. 2009;461:272–276. doi: 10.1038/nature08250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stenson PD, Mort M, Ball EV, Howells K, Phillips AD, Thomas NS, Cooper DN. The Human Gene Mutation Database: 2008 update. Genome Med. 2009;1:13. doi: 10.1186/gm13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Durbin RM, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hu J, Ng PC. Predicting the effects of frameshifting indels. Genome Biol. 2012;13:R9. doi: 10.1186/gb-2012-13-2-r9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sebro R, Levy H, Schneck K, Dimmock D, Raby BA, Cannon CL, Broeckel U, Risch NJ. Cystic fibrosis mutations for p.F508del compound heterozygotes predict sweat chloride levels and pancreatic sufficiency. Clinical Genetics. 2011 doi: 10.1111/j.1399-0004.2011.01804.x. October 28 (doi: 10.1111/j.1399-0004.2011.01804.x; epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang LL, Yang AK, Li Y, Liu JP, Zhou SF. Phenotype prediction of deleterious nonsynonymous single nucleotide polymorphisms in human alcohol metabolism-related genes: a bioinformatics study. Alcohol. 2010;44:425–438. doi: 10.1016/j.alcohol.2010.05.009. [DOI] [PubMed] [Google Scholar]
- 9.Yu ET, Hadi MZ. Bioinformatic processing to identify single nucleotide polymorphism that potentially affect Ape1 function. Mutat. Res. 2011;722:140–146. doi: 10.1016/j.mrgentox.2010.06.015. [DOI] [PubMed] [Google Scholar]
- 10.Lubbe SJ, Pittman AM, Matijssen C, Twiss P, Olver B, Lloyd A, Qureshi M, Brown N, Nye E, Stamp G, et al. Evaluation of germline BMP4 mutation as a cause of colorectal cancer. Hum. Mutat. 2011;32:E1928–E1938. doi: 10.1002/humu.21376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clague J, Wilhoite G, Adamson A, Bailis A, Weitzel JN, Neuhausen SL. RAD51C germline mutations in breast and ovarian cancer cases from high-risk families. PLoS One. 2011;6:e25632. doi: 10.1371/journal.pone.0025632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fan Y, Chen J, Wang W, Wu P, Zhi W, Xue B, Zhang W, Wang Y. Influence of eight unclassified missense variants of the MLH1 gene on Lynch syndrome susceptibility. Biochem. Genet. 2011;50:84–93. doi: 10.1007/s10528-011-9467-z. [DOI] [PubMed] [Google Scholar]
- 13.Ong SH, Yip JT, Chen YL, Liu W, Harun S, Lystiyaningsih E, Heriyanto B, Beckett CG, Mitchell WP, Hibberd ML, et al. Periodic re-emergence of endemic strains with strong epidemic potential-a proposed explanation for the 2004 Indonesian dengue epidemic. Infect. Genet. Evol. 2008;8:191–204. doi: 10.1016/j.meegid.2007.12.005. [DOI] [PubMed] [Google Scholar]
- 14.Till BJ, Reynolds SH, Weil C, Springer N, Burtner C, Young K, Bowers E, Codomo CA, Enns LC, Odden AR, et al. Discovery of induced point mutations in maize genes by TILLING. BMC Plant Biol. 2004;4:12. doi: 10.1186/1471-2229-4-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Till BJ, Cooper J, Tai TH, Colowit P, Greene EA, Henikoff S, Comai L. Discovery of chemically induced mutations in rice by TILLING. BMC Plant Biol. 2007;7:19. doi: 10.1186/1471-2229-7-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smits BM, van Zutphen BF, Plasterk RH, Cuppen E. Genetic variation in coding regions between and within commonly used inbred rat strains. Genome Res. 2004;14:1285–1290. doi: 10.1101/gr.2155004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guryev V, Berezikov E, Malik R, Plasterk RH, Cuppen E. Single nucleotide polymorphisms associated with rat expressed sequences. Genome Res. 2004;14:1438–1443. doi: 10.1101/gr.2154304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gharahkhani P, O'Leary CA, Kyaw-Tanner M, Sturm RA, Duffy DL. A non-synonymous mutation in the canine Pkd1 gene is associated with autosomal dominant polycystic kidney disease in Bull Terriers. PLoS One. 2011;6:e22455. doi: 10.1371/journal.pone.0022455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gunther T, Schmid KJ. Deleterious amino acid polymorphisms in Arabidopsis thaliana and rice. Theor. Appl. Genet. 2010;121:157–168. doi: 10.1007/s00122-010-1299-4. [DOI] [PubMed] [Google Scholar]
- 20.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ng PC, Henikoff S. Accounting for human polymorphisms predicted to affect protein function. Genome Res. 2002;12:436–446. doi: 10.1101/gr.212802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ng PC, Henikoff S. Predicting deleterious amino acid substitutions. Genome Res. 2001;11:863–874. doi: 10.1101/gr.176601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sunyaev S, Ramensky V, Bork P. Towards a structural basis of human non-synonymous single nucleotide polymorphisms. Trends Genet. 2000;16:198–200. doi: 10.1016/s0168-9525(00)01988-0. [DOI] [PubMed] [Google Scholar]
- 24.Thusberg J, Vihinen M. Pathogenic or not? And if so, then how? Studying the effects of missense mutations using bioinformatics methods. Hum. Mutat. 2009;30:703–714. doi: 10.1002/humu.20938. [DOI] [PubMed] [Google Scholar]
- 25.Thusberg J, Olatubosun A, Vihinen M. Performance of mutation pathogenicity prediction methods on missense variants. Hum. Mutat. 2011;32:358–368. doi: 10.1002/humu.21445. [DOI] [PubMed] [Google Scholar]
- 26.Li B, Krishnan VG, Mort ME, Xin F, Kamati KK, Cooper DN, Mooney SD, Radivojac P. Automated inference of molecular mechanisms of disease from amino acid substitutions. Bioinformatics. 2009;25:2744–2750. doi: 10.1093/bioinformatics/btp528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bao L, Zhou M, Cui Y. nsSNPAnalyzer: identifying disease-associated nonsynonymous single nucleotide polymorphisms. Nucleic Acids Res. 2005;33:W480–W482. doi: 10.1093/nar/gki372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee W, Zhang Y, Mukhyala K, Lazarus RA, Zhang Z. Bi-directional SIFT predicts a subset of activating mutations. PLoS One. 2009;4:e8311. doi: 10.1371/journal.pone.0008311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hicks S, Wheeler DA, Plon SE, Kimmel M. Prediction of missense mutation functionality depends on both the algorithm and sequence alignment employed. Hum. Mutat. 2011;32:661–668. doi: 10.1002/humu.21490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 31.Ng PC, Henikoff S. SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.International HapMap Consortium. The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 33.Stenson PD, Ball EV, Mort M, Phillips AD, Shiel JA, Thomas NS, Abeysinghe S, Krawczak M, Cooper DN. Human Gene Mutation Database (HGMD): 2003 update. Hum. Mutat. 2003;21:577–581. doi: 10.1002/humu.10212. [DOI] [PubMed] [Google Scholar]
- 34.Li K, Stockwell TB. VariantClassifier: a hierarchical variant classifier for annotated genomes. BMC Res. Notes. 2010;3:191. doi: 10.1186/1756-0500-3-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dreszer TR, Karolchik D, Zweig AS, Hinrichs AS, Raney BJ, Kuhn RM, Meyer LR, Wong M, Sloan CA, Rosenbloom KR, et al. The UCSC Genome Browser database: extensions and updates 2011. Nucleic Acids Res. 2012;40:D918–D923. doi: 10.1093/nar/gkr1055. [DOI] [PMC free article] [PubMed] [Google Scholar]