Abstract
jpHMM is a very accurate and widely used tool for recombination detection in genomic sequences of HIV-1. Here, we present an extension of jpHMM to analyze recombinations in viruses with circular genomes such as the hepatitis B virus (HBV). Sequence analysis of circular genomes is usually performed on linearized sequences using linear models. Since linear models are unable to model dependencies between nucleotides at the 5′- and 3′-end of a sequence, this can result in inaccurate predictions of recombination breakpoints and thus in incorrect classification of viruses with circular genomes. The proposed circular jpHMM takes into account the circularity of the genome and is not biased against recombination breakpoints close to the 5′- or 3′-end of the linearized version of the circular genome. It can be applied automatically to any query sequence without assuming a specific origin for the sequence coordinates. We apply the method to genomic sequences of HBV and visualize its output in a circular form. jpHMM is available online at http://jphmm.gobics.de for download and as a web server for HIV-1 and HBV sequences.
INTRODUCTION
Recombination analysis in viruses with circular genomes is usually performed with linear models on artificially linearized sequences of the circular genomes. When local dependencies, such as commonly modeled by hidden Markov models (HMM) or sliding window techniques, exist in a circular genome, these imply dependencies between the 5′- and 3′-end in the linearized version of the genome. Such dependencies are not modeled by linear approaches. As a consequence, recombination breakpoints located closely to the 5′- or 3′-end of the linearized sequence may be missed or erroneously predicted right at the origin for the sequence coordinates, if two different genotypes are predicted at both sequence ends. This can emphasize wrong recombination hotspots and lead to incorrect classification of circular viral genomes.
The hepatitis B virus (HBV) is such a virus with a circular genome. It is estimated that >2 billion people worldwide have been infected with HBV (1), among whom ∼360 million are chronically infected. Chronic hepatitis B infection can lead to serious illness, such as liver cirrhosis and hepatocellular carcinoma, as well as death. Eight different HBV ‘genotypes’, named alphabetically A-H, and several subgenotypes have been classified (2–7). Recombination among these (sub)genotypes is very common. The current classification system for HBV is based on sequence similarity (8) and recombinant forms are classified as (sub)genotypes. For recombination detection tools, a clear definition of pure genotypes is necessary to detect further recombinant forms of known genotypes. Thus, the selection of the parental genotype sequences must be well defined.
Three popular programs for recombination detection in HBV are Simplot (9), RDP3 (10) and TreeOrder Scan (11). All three use linear models, but two of them provide special features for circular genomes. Simplot provides a graph reflecting the similarity of a query sequence to a panel of reference sequences and predicts recombination breakpoints. RDP3 uses a range of recombination detection tools to identify recombinant sequences within a given set of aligned sequences. Besides the location of breakpoints, parental sequences of recombinants are determined among the given sequences. For circular genomes, recombination events that wrap around the sequence end are allowed and breakpoints at the sequence end are considered as real breakpoints. But, to our knowledge, the sequence end is classified independently from the beginning of the sequence and vice versa. TreeOrder Scan is part of the simple sequence editor (12). It uses several methods to evaluate the relationship between group membership and sequence order in phylogenetic trees generated from their nucleotide sequences. Positions in an alignment of these sequences where phylogeny relationship change, e.g. as a result of recombination, are visualized. Dragging or moving sequences in a circular alignment allows nucleotides to be taken from the end of the alignment to the beginning, or vice versa. However, it is not clear how this manual editing influences the result.
Here, we present an extension of our ‘jumping profile Hidden Markov Model’ (jpHMM) (13–15) for recombination detection in circular viral genomes. jpHMM was previously developed to detect recombinations in genomic sequences of HIV-1. Evaluation on simulated recombined sequences as well as real viral genomes showed that it is one of the most accurate methods to predict recombination breakpoints in HIV-1 genomes. The proposed circular jpHMM approach inherently detects recombination breakpoints in circular genomes, taking into account dependencies between nucleotides at both ends of the linearized version of a circular genome. We apply the circular jpHMM to detect recombinations in HBV genomes.
MATERIALS AND METHODS
jpHMM
jpHMM is a probabilistic model that we developed to compare single nucleotide sequences to a given multiple alignment of a sequence family (13). Given a partition of the alignment into subclasses, called ‘subtypes’, each subtype is modeled as a profile HMM (16). In addition to the usual state transitions ‘within’ a profile HMM, transitions, called ‘jumps’, ‘between’ the different profile HMMs are allowed. To these jumps, a ‘jump probability’ is assigned. The alignment of a query sequence to the given multiple alignment is then defined by the most probable path through the model generating the sequence, the so-called ‘Viterbi path’ (17), allowing jumps between different subtypes. This alignment is called the ‘jumping alignment’ of the query sequence to the given alignment (18). Positions of jumps between different subtypes define recombination breakpoints. Additionally, an ‘interval’ estimate for each predicted breakpoint (‘breakpoint interval’) and a tagging of regions in which the model is uncertain about the predicted subtype (‘uncertainty regions’) are determined (15).
A jpHMM for circular genomes
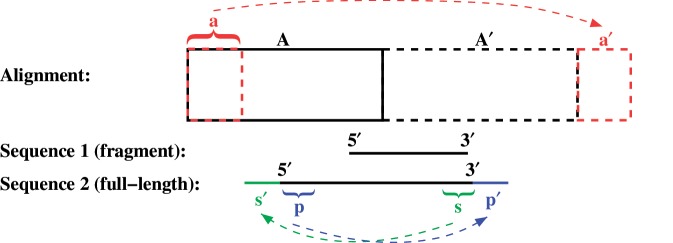
To allow an accurate recombination prediction at the 5′- and 3′-end of the linearized version of circular genomes, each full-length input sequence is extended at both sequence ends: The prefix (5′-) and the suffix (3′) of the sequence are copied and concatenated to the original 3′- and 5′-end respectively (Figure 1). Thus, possible dependencies between nucleotides at the 5′- and 3′-end of the sequence are considered in the recombination prediction. Also, nearly full-length but not complete genomes, where some linkage between both sequence ends can be expected as well, are extended in this way. In these sequences, the missing part is modeled by delete states.
Figure 1.
The input alignment A (roughly sketched by the black rectangle) is duplicated (A′) and a prefix (a) is copied and concatenated to the end of the alignment (a′). Each nearly full-length sequence is extended by copying and concatenating the prefix (p) as well as the suffix (s) of the sequence to its 3′ end (p′) and 5′ end (s′), respectively.
To enable an alignment of extended, (nearly) full-length as well as of fragmental sequences to the input alignment, regardless of the chosen origin for the sequence coordinates, the alignment is extended as well (Figure 1). It is duplicated and a prefix is copied and concatenated to the end of the alignment. On the basis of this extended alignment, the model is built.
Since most extended as well as fragmental sequences can be aligned nearly completely to two different regions in the extended alignment, the direct application of jpHMM would result in an unnecessary waste of runtime and memory. For this reason, at first, the location of each (extended) query sequence with respect to the extended alignment is determined. The sequence is aligned to the sequences in the alignment using the BLAST-like alignment tool BLAT (19). For each sequence position, these pairwise alignments define an interval of alignment columns to which the respective sequence position is allowed to be aligned with jpHMM. Only states corresponding to the respective interval of alignment columns are allowed to generate a certain nucleotide of the query sequence. This reduces the search space of the Viterbi path to only a small number of states for each sequence position. Thus, extended full-length as well as fragmental sequences can be analyzed quickly without assuming a specific origin for the sequence coordinates. The average runtime of the circular jpHMM is 48 s for a full-length HBV sequence. (We also applied this pre-alignment version to HIV genomes, which are linear, and could reduce the jpHMM runtime by half, i.e. from 7 min 18 s to 3 min 26 s.)
The predicted recombination is visualized in a circular form (Figure 2) using the software package Circos (20). For extended, full-length sequences, the output only comprises the prediction for the original sequence. Different subtypes at the 5′- and 3′-end of the sequence imply a breakpoint at this position. For a consistent representation of sequence positions, all sequence position numbers are given relative to a chosen reference strain.
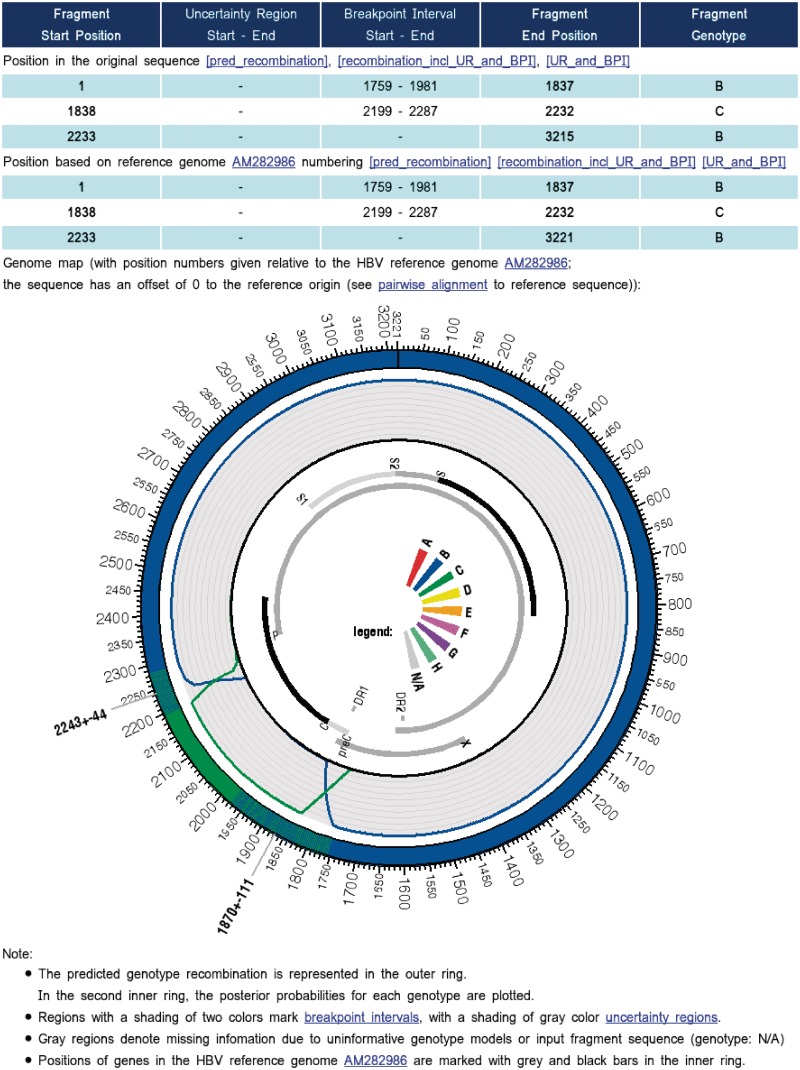
Figure 2.
Extract of the jpHMM web server output for a real HBV/BC recombinant. The output contains a list of fragments from the input sequence that are assigned to different HBV genotypes, including breakpoint intervals and uncertainty regions. The predicted recombination is represented graphically in a circular form using the software package Circos (20). Regions with a shading of two colors mark breakpoint intervals, e.g. region 2243±44 (outer ring). The posterior probabilities for each genotype are plotted in the second inner ring. All sequence position numbers are given relative to the HBV reference genome AM282986. Positions of genes in the genome are marked with gray and black bars (inner ring). ‘N/A’ in the color legend (middle) denotes for ‘not assigned’.
Application to HBV
For the application of the circular jpHMM to recombination detection in HBV, we chose a length of 500 nt for the extension of a nearly full-length (>3000 nt) query sequence at both sequence ends. The parental genotype sequences for the input alignment have been carefully selected. On the basis of a small alignment with well-defined pure genotype sequences, all full-length HBV sequences published in GenBank (21) in December 2009 were tested for recombination with jpHMM using a very high jump probability of 10−3. All confirmed pure genotype sequences were clustered with CD-HIT-EST (22) using different sequence identity thresholds to obtain about 50 (if available) representative sequences for each genotype. The resulting 339 sequences were aligned with Muscle (23). The input alignment is part of the jpHMM source code archive and can be downloaded.
Due to the lack of real recombinant sequences with exactly known breakpoint positions, the jump probability jp and the pseudo count α for the emission probabilities of the model were estimated jointly on 276 semi-artificial recombinant HBV sequences with artificially introduced breakpoints. The training sequences were created as described in the section ‘Evaluation’, but the number (0–4) and the position of the breakpoints in the genome were chosen randomly for each sequence. Several criteria such as the accuracy of the predicted breakpoint intervals or the predicted set of parental genotypes (see ‘Results’ section) were used to estimate the optimal pair of parameters ( jp, α). The resulting jump probability is 10−7 and as pseudo count for the emission probabilities we use α = 0.009.
Web server
jpHMM is available online at http://jphmm.gobics.de/. The user can paste or upload up to 20 full-length HBV genomic sequences or fragments at a time in FASTA format. A hyperlink to the results of the program run, which are stored on the server for 7 days, is given and can be bookmarked. If the user enters an Email address, this hyperlink will also be returned to the user by Email. The result contains for each sequence the predicted recombination, including uncertainty regions and breakpoint intervals, in text format. Additionally, a graphical representation of the predicted recombinant fragments within the HBV genome is given in a circular form. This plot also contains the posterior probabilities of the genotypes for each sequence position. All result files can be downloaded. Figure 2 shows an excerpt of the jpHMM output for a real HBV/BC recombinant sequence.
For the definition of uncertainty regions and breakpoint intervals, we use a threshold of 0.99 for the posterior probabilities (15). As reference genome, we chose the well-annotated sequence AM282986 (24), which belongs to genotype A. This sequence is also part of the multiple sequence alignment we use to build the model. The alignment of each input sequence to the reference genome, defined by jpHMM, is provided for download.
Evaluation
The accuracy of the circular jpHMM was evaluated on three data sets (DS1-3), each comprising 280 semi-artificial recombinant HBV sequences. As customary, each test sequence is a recombinant of two ‘real-world’ sequences from two different HBV genotypes (A–H) with breakpoints artificially introduced at certain positions: in DS1, a breakpoint is introduced at every 1000th position in the sequence, in DS2 alternately at every 500th and 1500th position and in DS3 alternately at every 300th and 1500th position, numbering according to genome AM282986. For example, in sequences of DS2, alternating short segments (length 500 nt) of one genotype are interrupted by long segments (length 1500 nt) from another genotype. All test sequences contain four recombination breakpoints. To simulate previously unobserved sequences, the two parental sequences of each test sequence are removed from the multiple alignment that is used to build the model in the respective program run. The test data sets can be downloaded from http://jphmm.gobics.de/download.
The accuracy of the predicted recombination was evaluated in terms of the accuracy of the predicted breakpoint intervals (BPIs), the accuracy of the predicted recombination pattern and the accuracy of the predicted parental genotypes at each sequence position.
RESULTS
The accuracy of the predicted breakpoint intervals was assessed by their ‘specificity’, i.e. the number of breakpoint intervals that contain a true breakpoint, and their ‘sensitivity’, i.e. the number of breakpoints that were detected with the predicted breakpoint intervals. A breakpoint is defined as ‘detected’ if it is located in a predicted breakpoint interval and if the two neighboring genotypes are predicted correctly.
Breakpoint intervals and uncertainty regions have been defined on the basis of the posterior probabilities for different thresholds. In Table 1, the accuracy of the predicted breakpoint intervals and parental genotypes is given for different thresholds for data set DS1. As default threshold, we chose 0.99, since it provides the best trade-off between the average length and the accuracy of the predicted breakpoint intervals. For this threshold, a specificity as well as a sensitivity of 99.64% was observed for DS1 (Table 1, Columns 2 and 3), with an average breakpoint interval length of 21 nt. In DS2, the specificity was equal to the sensitivity too, namely 99.46%, with an average breakpoint interval length of 26 nt. The number of breakpoints was predicted correctly in all sequences of both data sets, the breakpoints that could not be detected are located outside of the predicted breakpoint intervals. As it can be seen in Table 1, increasing the posterior probability threshold to 0.9999, and thus enlarging the predicted breakpoint intervals, leads to a specificity and a sensitivity of 100%. This also holds for DS2.
Table 1.
Accuracy of the predicted BPIs and parental genotypes for different posterior probability thresholds for data set DS1
| Threshold | BPI |
BPI length |
Positions ∉{UR/BPI} | ||
|---|---|---|---|---|---|
| Spec. (%) | Sens. (%) | Average | Min./ Max. | classified correctly (%) | |
| 0.90 | 88.98 | 86.52 | 14.16 | 2/87 | 99.94 |
| 0.95 | 95.32 | 94.46 | 16.65 | 2/124 | 99.97 |
| 0.99 | 99.64 | 99.64 | 20.85 | 1/240 | 100 |
| 0.9999 | 100 | 100 | 32.62 | 2/555 | 100 |
In Column 1, the threshold for the posterior probabilities is given. In Columns 2 and 3, the specificity (Spec.) and the sensitivity (Sens.) of the predicted BPIs defined by this threshold are given. The average and the minimal and maximal length of these BPIs are given in Columns 4 and 5. Column 6 shows the percentage of sequence positions located outside of uncertainty regions (URs) and BIPs that are classified correctly.
In DS3, a specificity of 98.47% (average breakpoint interval length of 36 nt) was achieved, which can also be increased to 100% using a posterior probability threshold of 0.9999. In contrast to DS1 and DS2, the sensitivity of the predicted breakpoint interval is lower than the specificity, namely 97.77%. The reason is that in four D/E recombinants, one short segment of genotype D (300 nt) was not identified. For all other sequences studied in this article, the recombination pattern, i.e. the sequence of subtypes, was predicted correctly.
The accuracy of predicted genotypes was assessed by the number of sequence positions assigned to the correct genotype. In DS1, 99.32% of the sequence positions were classified correctly, 99.29% in DS2 and 98.19% in DS3. Considering only sequence positions located outside of predicted breakpoint interval and uncertainty regions, in DS1 and DS2, even 100% (rounded to two decimal places) of the positions were classified correctly. In DS3, 0.17% of the sequence positions outside of uncertainty regions and breakpoint intervals were classified incorrectly, which corresponds to only 5.4 nt in a sequence of length 3200.
CONCLUSION
The proposed circular jpHMM approach predicts recombinations in a circular viral genome automatically without assuming a specific origin for the sequence coordinates. No manual editing of the sequences is required. By the extension of the query sequence at both sequence ends, dependencies between the 5′- and 3′-end of the linearized version of the circular genome are taken into account and the method is not biased against recombination breakpoints close to the chosen 5′- or 3′-end of the linearized sequence. The high accuracy of the recombination prediction for semi-artificial HBV recombinants demonstrates that jpHMM is a suitable and powerful tool for recombination detection in HBV genomes. Researchers will also benefit from the circular representation of the predicted recombination.
FUNDING
The Deutsche Forschungsgemeinschaft [STA 1009/5-1 to M.S.]; ANRS and InVS (to M.A.C.). Funding for open access charge: Department of Bioinformatics, Georg-August-Universität Göttingen, Germany.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank the anonymous referees for their suggestions.
REFERENCES
- 1.WHO. Hepatitis B Vaccines. Wkly Epidemiol Rec. 2009;84:405–420. [PubMed] [Google Scholar]
- 2.Okamoto H, Tsuda F, Sakugawa H, Sastrosoewignjo RI, Imai M, Miyakawa Y, Mayumi M. Typing hepatitis B virus by homology in nucleotide sequence: comparison of surface antigen subtypes. J. Gen. Virol. 1988;69:2575–2583. doi: 10.1099/0022-1317-69-10-2575. [DOI] [PubMed] [Google Scholar]
- 3.Naumann H, Schaefer S, Yoshida CFT, Gaspar AMC, Repp R, Gerlich WH. Identification of a new hepatitis B virus (HBV) genotype from Brazil that expresses HBV surface antigen subtype adw4. J. Gen. Virol. 1993;74:1627–1632. doi: 10.1099/0022-1317-74-8-1627. [DOI] [PubMed] [Google Scholar]
- 4.Norder H, Hammas B, Löfdahl S, Couroucé A-M, Magnius LO. Comparison of the amino acid sequences of nine different serotypes of hepatitis B surface antigen and genomic classification of the corresponding hepatitis B virus strains. J. Gen. Virol. 1992;73:1201–1208. doi: 10.1099/0022-1317-73-5-1201. [DOI] [PubMed] [Google Scholar]
- 5.Norder H, Couroucé A-M, Magnius LO. Complete genomes, phylogenetic relatedness, and structural proteins of six strains of the hepatitis B virus, four of which represent two new genotypes. Virology. 1994;198:489–503. doi: 10.1006/viro.1994.1060. [DOI] [PubMed] [Google Scholar]
- 6.Stuyver L, De Gendt S, Van Geyt C, Zoulim F, Fried M, Schinazi RF, Rossau R. A new genotype of hepatitis B virus: complete genome and phylogenetic relatedness. J. Gen. Virol. 2000;81:67–74. doi: 10.1099/0022-1317-81-1-67. [DOI] [PubMed] [Google Scholar]
- 7.Arauz-Ruiz P, Norder H, Robertson BH, Magnius LO. Genotype H: a new Amerindian genotype of hepatitis B virus revealed in Central America. J. Gen. Virol. 2002;83:2059–2073. doi: 10.1099/0022-1317-83-8-2059. [DOI] [PubMed] [Google Scholar]
- 8.Kramvis A, Arakawa K, Yu MC, Nogueira R, Stram DO, Kew MC. Relationship of serological subtype, basic core promoter and precore mutations to genotypes/subgenotypes of hepatitis B virus. J. Med. Virol. 2008;80:27–46. doi: 10.1002/jmv.21049. [DOI] [PubMed] [Google Scholar]
- 9.Lole KS, Bollinger RC, Paranjape RS, Gadkari D, Kulkarni SS, Novak NG, Ingersoll R, Sheppard HW, Ray SC. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J. Virol. 1999;73:152–160. doi: 10.1128/jvi.73.1.152-160.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martin DP, Lemey P, Lott M, Moulton V, Posada D, Lefeuvre P. RDP3: a flexible and fast computer program for analyzing recombination. Bioinformatics. 2010;26:2462–2463. doi: 10.1093/bioinformatics/btq467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Simmonds P, Midgley S. Recombination in the genesis and evolution of hepatitis B virus genotypes. J. Virol. 2005;79:15467–15476. doi: 10.1128/JVI.79.24.15467-15476.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Simmonds P. SSE: a nucleotide and amino acid sequence analysis platform. BMC Res. Notes. 2012;5:50. doi: 10.1186/1756-0500-5-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schultz A-K, Zhang M, Leitner T, Kuiken C, Korber B, Morgenstern B, Stanke M. A jumping profile Hidden Markov Model and applications to recombination sites in HIV and HCV genomes. BMC Bioinformatics. 2006;7:265. doi: 10.1186/1471-2105-7-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang M, Schultz A-K, Calef C, Kuiken C, Leitner T, Korber B, Morgenstern B, Stanke M. jpHMM at GOBICS: a web server to detect genomic recombinations in HIV-1. Nucleic Acids Res. 2006;34:W463–W465. doi: 10.1093/nar/gkl255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schultz A-K, Zhang M, Bulla I, Leitner T, Korber B, Morgenstern B, Stanke M. jpHMM: improving the reliability of recombination prediction in HIV-1. Nucleic Acids Res. 2009;37:W647–W651. doi: 10.1093/nar/gkp371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eddy SR. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- 17.Viterbi A. Error bounds for convolutional codes and an asymptotically optimum decoding algorithm. IEEE Trans. Inf. Theory. 1967;13:260–269. [Google Scholar]
- 18.Spang R, Rehmsmeier M, Stoye J. A novel approach to remote homology detection: jumping alignments. J. Comp. Biol. 2002;9:747–760. doi: 10.1089/106652702761034172. [DOI] [PubMed] [Google Scholar]
- 19.Kent WJ. BLAT - The BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krzywinski M, Schein J, Birol İ, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. Circos: An information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bilofsky HS, Burks C, Fickett JW, Goad W, Lewitter FI, Rindone WP, Swindell CD, Tung CS. The GenBank genetic sequence databank. Nucleic Acids Res. 1986;14:1–4. doi: 10.1093/nar/14.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li W, Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 23.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic. Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Panjaworayan N, Roessner S, Firth A, Brown C. HBVRegDB: annotation, comparison, detection and visualization of regulatory elements in hepatitis B virus sequences. J. Virol. 2007;4:136. doi: 10.1186/1743-422X-4-136. [DOI] [PMC free article] [PubMed] [Google Scholar]