Abstract
RNA secondary structure is required for the proper regulation of the cellular transcriptome. This is because the functionality, processing, localization and stability of RNAs are all dependent on the folding of these molecules into intricate structures through specific base pairing interactions encoded in their primary nucleotide sequences. Thus, as the number of RNA sequencing (RNA-seq) data sets and the variety of protocols for this technology grow rapidly, it is becoming increasingly pertinent to develop tools that can analyze and visualize this sequence data in the context of RNA secondary structure. Here, we present Sequencing Annotation and Visualization of RNA structures (SAVoR), a web server, which seamlessly links RNA structure predictions with sequencing data and genomic annotations to produce highly informative and annotated models of RNA secondary structure. SAVoR accepts read alignment data from RNA-seq experiments and computes a series of per-base values such as read abundance and sequence variant frequency. These values can then be visualized on a customizable secondary structure model. SAVoR is freely available at http://tesla.pcbi.upenn.edu/savor.
INTRODUCTION
The secondary structure of an RNA molecule comprises specific base pairing interactions encoded within the primary nucleotide sequence. The formation of secondary structure is vital to the maturation and function of many classes of RNAs. For example, the classic clover-leaf folding pattern of tRNAs is necessary for their function in translation, while the processing of multiple classes of small regulatory RNAs requires formation of specific secondary structures (1,2). Recently, the advent of high-throughput RNA sequencing (RNA-seq) has enabled unbiased, genome-wide studies of many RNA populations within the cell. RNA-seq and its variant protocols have been recently used to study a wide range of biological phenomena, including RNA silencing (3,4), RNA–protein interactions (5,6) and protein translation (7), to name a few. These experiments, along with several recent studies of RNA base pairing (4,8–10), have highlighted the functional significance of RNA structure on a global scale.
Although the importance of RNA secondary structure is clear, most existing tools for RNA-seq analysis, such as DESeq (11), Myrna (12), Cufflinks (13), and Galaxy (14), primarily report RNA-seq analyses in the context of linear transcript models and do not support a structure-based interpretation. On the other hand, tools that do enable visualization and annotation of RNA structure models [e.g. RNAstructure (15), RNAfold (16), etc.] are focused on the problem of RNA secondary structure prediction and are not easily applicable to analysis of RNA-seq data. To address this gap, we have developed Sequencing Annotation and Visualization of RNA structures (SAVoR), which neatly integrates common RNA-seq analyses with a structure-based annotation and visualization framework (Figure 1). To do this, SAVoR extracts sequencing data from user-specified RNA-seq alignment files and computes a series of per-nucleotide values such as read abundance and sequence variant frequency, which are then directly plotted on a customizable structural model. The entire process is streamlined via a simple web interface and is completely platform independent. The uses of SAVoR range from a quick look at a transcript of interest to fully customized and annotated publication quality structure models.
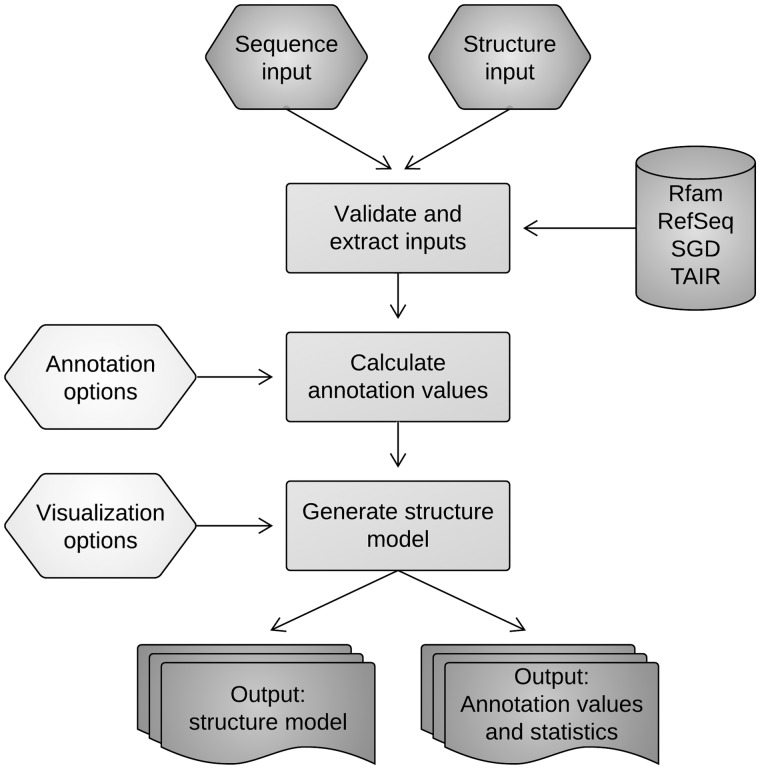
Figure 1.
The SAVoR workflow. Upon validation of user input, the primary sequence and genomic location of the user-submitted transcript(s) are determined, and intersecting sequence reads are converted to the desired annotation values. The secondary structure is then determined and plotted with the specified visualization options.
SAVOR WEB SERVER
Input
SAVoR requires the user to enter an RNA transcript sequence and a secondary structure as input. The sequence can be entered as a primary sequence in plain text or FASTA format, an Rfam (17) or transcript [Refseq (18), SGD (19) or TAIR (20)] accession number, or the genomic location by chromosomal range and strand information (Figure 2). Currently, the input sequence is restricted to 20 000 nt in length. If an Rfam accession number is entered and multiple matching entries are located (often the case for repetitive RNA elements such as tRNAs), then SAVoR lists all matching entries from which the user can then select the desired locus. If a primary sequence is entered and any genome-based annotation is selected, then BLASTN (21) with ‘-gapopen 999 gapextend 999’ and otherwise default parameters will be used to determine the genomic location of the input sequence. The user will be prompted to select from a list of the top 20 BLASTN results that pass an E-value cutoff of 1e−3. The user can use a simple drop-down menu to select the reference genome, which is used by SAVoR to retrieve database entries and primary sequence data. SAVoR currently supports the latest reference genome releases for human, mouse, Drosophila melanogaster (fruit fly), Saccharomyces cerevisiae (budding yeast), Arabidopsis thaliana and Caenorhabditis elegans and contains 3831 Rfam entries and 167 157 RefSeq/SGD/TAIR entries.
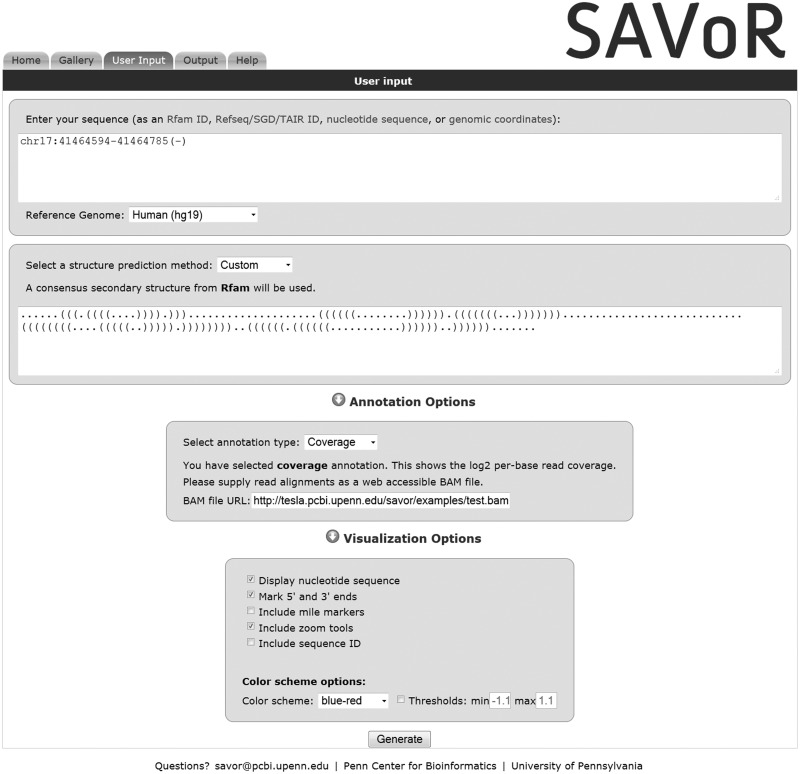
Figure 2.
SAVoR can be used to visualize RNA-seq results in the context of predicted RNA secondary structures. In this example, the ‘User Input’ tab of the SAVoR web interface is populated with a sample input. The desired transcript is specified by UCSC-style genomic coordinates, and a custom structure is provided in dot-parenthesis notation. The ‘coverage’ annotation type has been selected and the URL for a web-accessible BAM file of read alignments provided. All optional visualization settings have been selected, and a blue-red color scheme will be used.
Specifying the secondary structure
Depending on the type of input sequence and RNA-seq data, the user has four options to specify how the model of RNA secondary structure is generated. For example, SAVoR can retrieve the secondary structure from the Rfam database when the input is an Rfam accession ID. Additionally, the RNAfold program can be used with or without experimental constraints to fold the sequence into its minimum free energy state. If the constrained option is selected, the log2 abundance ratios of structure-informative RNA-seq data sets (4,8–10) are used to derive experimental constraints for structure prediction. Specifically, in the resulting structure model, a base will be paired when the dsRNA-seq to ssRNA-seq abundance ratio for that nucleotide exceeds some given threshold and vice versa (4,9). SAVoR will then use RNAfold to find the best secondary structure model based on the given constraints. Finally, the user can enter a specific secondary structure using the common dot-parenthesis notation (22).
Generating per-base annotations
Next, the user specifies the type of annotation to be overlaid on the RNA secondary structure model. Importantly, SAVoR supports remote access to indexed BAM files, which are highly compact files that contain read alignments from an RNA-seq experiment. SAVoR directly extracts sequencing reads that intersect with the input RNA transcript without requiring the user to upload the entire BAM file. Extracted reads are then converted to per-nucleotide annotation values. SAVoR can generate four different annotation types based on BAM files from RNA-seq or other types of high-throughput sequencing experiments: (i) read abundance (number of reads that cover each nucleotide base), (ii) endpoint abundance (number of reads whose 5′ or 3′ endpoint occurs at each nucleotide base), (iii) per-base mismatch frequency and (iv) per-base normalized log2 abundance ratio (for this analysis, the user is required to enter URL of two BAM files, which will be used by SAVoR to compute ratios).
It is worth noting that when log2 abundance or abundance ratio is selected, pseudo counts (adding 1 to the count of every position) are used to avoid numerical errors.
Alternatively, the user can upload a text file of custom annotation values using the UCSC Genome Browser BED format, a flexible tabular file format for genomic locations and associated data (http://genome.ucsc.edu/FAQ/FAQformat.html#format1). Currently, the BED-format file is limited to 5 Mb in size. Finally, the user can select from a series of visualization options that specify the markup and color scheme of the output structural model (these options are described in detail on the SAVoR website). Figure 2 shows an example input page that uses genomic coordinates for sequence input, a custom dot-parenthesis structure, and read coverage annotation with default visualization settings. The user can try out this sample input by clicking the ‘Sample input’ link on the SAVoR home page.
Output
After the user submits the input data, the ‘Output’ tab is automatically displayed (Figure 3). Progress indicators are shown for each step of the SAVoR workflow, along with warnings at steps that may require additional processing time. SAVoR validates all user-supplied data (e.g. that the input sequence only contains valid nucleotide characters) and reports any detected anomalies to help the user fix any errors in the input. Upon completion, the output structural model is directly displayed in the web browser, along with the calculated annotation values in tabular format. A legend showing the color scheme and annotation type is displayed in the top left corner of the output model. The sequence and structure are displayed using the default layout by RNAplot (16) with annotation values overlaid. The 5′ and 3′ ends of the transcript, as well as the position of every 10th nucleotide, are marked to facilitate location of a specific region of interest. The entire model can be scaled and panned as desired using standard browser tools.
Figure 3.
SAVoR produces highly informative, user specified, annotated models of RNA secondary structure. The ‘Output’ tab of the SAVoR web interface is dynamically rendered to show progress indicators and error messages. Each step of the pipeline is indicated, and a ‘Complete’ message appears upon successful completion. When the entire process is finished, the output (an annotated model of RNA secondary structure) is displayed, and links for downloading the output files are provided. Additionally, the calculated per-base annotation values are displayed in tabular form and can be downloaded as a plain text file.
Links to the output structure model in SVG, PDF, and PNG formats, and the annotation values in plain text format are provided as well. Importantly, files generated by each user submission are uniquely named and can only be viewed via these output links. The results are kept by the server for at least 72 h. If changes to the input data are desired, the user can simply click on the ‘User Input’ tab and directly modify the stored input values. While resubmission of the input form will result in rerunning of the entire SAVoR pipeline and generation of new output files, a typical SAVoR run requires <30 s for a 1 kb sequence.
Example uses
While we have streamlined the workflow design to strengthen its accessibility, SAVoR is also very flexible. We describe three example use cases to illustrate this point. Corresponding figures can be found in the Gallery page on the SAVoR website.
Visualizing read distribution across a known transcript: The user specifies an Rfam or RefSeq ID, the ‘coverage’ annotation option, and read alignments as a BAM file. This type of model can be used to look for biases in read distribution such as those derived from small RNAs produced from a precursor transcript.
Comparing experimental and computational base pairing predictions: The user specifies the ‘RNAfold’ structure prediction method and ‘log-ratio’ annotation option, and provides two BAM files (containing structure-informative RNA-seq data) as input. The resulting output can be used to compare base pairing predictions from a free-energy based computational approach (RNAfold) with experimentally derived base pairing data (log-ratio).
Visualizing single nucleotide polymorphisms (SNPs): The user uploads a UCSC Genome Browser BED format file of customized per-nucleotide values along with any set of sequence and structure inputs. For example, we can upload a file-containing SNP coordinates and use this to color known SNPs on the secondary structure; this allows the user to examine if population diversity correlates with predicted or experimentally determined RNA structural constraints.
Implementation
The SAVoR web server runs Apache 2.2.3 on a CentOS 5.7 machine with 2× Intel Xeon E5450 3.00 GHz processors and 16 GB RAM. Asynchronous JavaScript and XML (AJAX) technology is used to dynamically render PHP output into formatted HTML. A local MySQL database is used to store Rfam and Refseq/SGD/TAIR entries, and a local installation of BLAST+ is used to retrieve sequence and genomic locus information. Structure prediction is optionally performed using a local installation of RNAfold (version 1.8.4), and backbone layout is done using RNAplot. SAMtools (23) is used to extract annotation values from BAM files, and custom Perl and Ruby scripts are used to process BED files. Inkscape (version 0.47) is used to convert from the native SVG format to publication quality PDF and PNG output files. SAVoR has been tested extensively, and the internal programs were used to generate annotated models of RNA secondary structure in our recent publications (4,9).
CONCLUSIONS
The incredible power and versatility of high-throughput RNA-seq approaches have spurred many insights into RNA function, biogenesis, and structure, and offer almost endless possibilities for future studies of RNA biology (24–27). Interpretation of these data is fast becoming a bottleneck, and substantial efforts to aid in this process are currently necessary. With SAVoR, we have developed a unique and user-friendly interface to streamline the interpretation of RNA-seq data in the context of RNA secondary structure. Specifically, our web server directly computes per-nucleotide quantities from RNA-seq data sets and overlays these annotation values on a structural model. The uses of this web-based tool range from quick checks of data quality to production of fully customized publication quality figures, and will aid researchers in many aspects of RNA-seq analysis. We plan to extend SAVoR to directly retrieve annotations from the UCSC Genome Browser and other public genomic databases, thereby removing the need for users to generate their own annotation files and improving accessibility to different data sources beyond sequencing alignments. We also plan to add other methods for structure prediction and visualization such as conservation-informed prediction (28,29), and implement other RNA secondary structure layout options including circular and linear structure plots. These additional features will further aid interpretation of genome-scale data in the context of RNA secondary structure.
FUNDING
Funding for open access charge: Department of Pathology and Laboratory Medicine, University of Pennsylvania Perelman School of Medicine. This work was supported by National Science Foundation [MCB-1053846 to B.D.G.]; Penn Genome Frontiers Institute [pilot award to B.D.G. and L.S.W.]; National Institutes of Health [NIA AG10124 to B.D.G.and L.S.W.; NHGRI 5T32HG000046-13 to F.L. and P.R.]; SmithKline Beecham Center of Excellence in Geriatric Medicine through the Penn Institute on Aging [to M.C.].
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank members of Gregory and Wang labs, Mingyao Li, John Hogenesch, Chris Stoeckert, and other participants of the HTS working group at Penn for their constructive comments and suggestions.
REFERENCES
- 1.Cruz JA, Westhof E. The dynamic landscapes of RNA architecture. Cell. 2009;136:604–609. doi: 10.1016/j.cell.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 2.Sharp PA. The centrality of RNA. Cell. 2009;136:577–580. doi: 10.1016/j.cell.2009.02.007. [DOI] [PubMed] [Google Scholar]
- 3.Bracken CP, Szubert JM, Mercer TR, Dinger ME, Thomson DW, Mattick JS, Michael MZ, Goodall GJ. Global analysis of the mammalian RNA degradome reveals widespread miRNA-dependent and miRNA-independent endonucleolytic cleavage. Nucleic Acids Res. 2011;39:5658–5668. doi: 10.1093/nar/gkr110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zheng Q, Ryvkin P, Li F, Dragomir I, Valladares O, Yang J, Cao K, Wang LS, Gregory BD. Genome-wide double-stranded RNA sequencing reveals the functional significance of base-paired RNAs in Arabidopsis. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1001141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lebedeva S, Jens M, Theil K, Schwanhäusser B, Selbach M, Landthaler M, Rajewsky N. Transcriptome-wide analysis of regulatory interactions of the RNA-binding protein HuR. Mol. Cell. 2011;43:340–352. doi: 10.1016/j.molcel.2011.06.008. [DOI] [PubMed] [Google Scholar]
- 6.Mukherjee N, Corcoran DL, Nusbaum JD, Reid DW, Georgiev S, Hafner M, Ascano M, Jr, Tuschl T, Ohler U, Keene JD. Integrative regulatory mapping indicates that the RNA-binding protein HuR couples pre-mRNA processing and mRNA stability. Mol. Cell. 2011;43:327–339. doi: 10.1016/j.molcel.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kertesz M, Wan Y, Mazor E, Rinn JL, Nutter RC, Chang HY, Segal E. Genome-wide measurement of RNA secondary structure in yeast. Nature. 467:103–107. doi: 10.1038/nature09322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li F, Zheng Q, Ryvkin P, Dragomir I, Desai Y, Aiyer S, Valladares O, Yang J, Bambina S, Sabin LR, et al. Global analysis of RNA secondary structure in two metazoans. Cell Rep. 2012;1:69–82. doi: 10.1016/j.celrep.2011.10.002. [DOI] [PubMed] [Google Scholar]
- 10.Underwood JG, Uzilov AV, Katzman S, Onodera CS, Mainzer JE, Mathews DH, Lowe TM, Salama SR, Haussler D. FragSeq: transcriptome-wide RNA structure probing using high-throughput sequencing. Nat. Methods. 2010;7:995–1001. doi: 10.1038/nmeth.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Langmead B, Hansen KD, Leek JT. Cloud-scale RNA-sequencing differential expression analysis with Myrna. Genome Biol. 2010;11:R83. doi: 10.1186/gb-2010-11-8-r83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goecks J, Nekrutenko A, Taylor J. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 2010;11:R86. doi: 10.1186/gb-2010-11-8-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reuter JS, Mathews DH. RNAstructure: software for RNA secondary structure prediction and analysis. BMC Bioinformatics. 2010;11:129. doi: 10.1186/1471-2105-11-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hofacker IL, Stadler PF. Memory efficient folding algorithms for circular RNA secondary structures. Bioinformatics. 2006;22:1172–1176. doi: 10.1093/bioinformatics/btl023. [DOI] [PubMed] [Google Scholar]
- 17.Gardner PP, Daub J, Tate J, Moore BL, Osuch IH, Griffiths-Jones S, Finn RD, Nawrocki EP, Kolbe DL, Eddy SR, et al. Rfam: Wikipedia, clans and the "decimal" release. Nucleic Acids Res. 2011;39:D141–D145. doi: 10.1093/nar/gkq1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pruitt KD, Tatusova T, Brown GR, Maglott DR. NCBI Reference Sequences (RefSeq): current status, new features and genome annotation policy. Nucleic Acids Res. 2012;40:D130–D135. doi: 10.1093/nar/gkr1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cherry JM, Hong EL, Amundsen C, Balakrishnan R, Binkley G, Chan ET, Christie KR, Costanzo MC, Dwight SS, Engel SR, et al. Saccharomyces Genome Database: the genomics resource of budding yeast. Nucleic Acids Res. 2012;40:D700–D705. doi: 10.1093/nar/gkr1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lamesch P, Berardini TZ, Li D, Swarbreck D, Wilks C, Sasidharan R, Muller R, Dreher K, Alexander DL, Garcia-Hernandez M, et al. The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res. 2012;40:D1202–D1210. doi: 10.1093/nar/gkr1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hofacker IL, Fontana W, Stadler PF, Bonhoeffer LS, Tacker M, Schuster P. Fast folding and comparison of RNA secondary structures. Monatshefte für Chemie/Chemical Monthly. 1994;125:167–188. [Google Scholar]
- 23.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ozsolak F, Milos PM. RNA sequencing: advances, challenges and opportunities. Nat. Rev. Genet. 2011;12:87–98. doi: 10.1038/nrg2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Westhof E, Romby P. The RNA structurome: high-throughput probing. Nat. Methods. 2010;7:965–967. doi: 10.1038/nmeth1210-965. [DOI] [PubMed] [Google Scholar]
- 26.Schmitz RJ, Zhang X. High-throughput approaches for plant epigenomic studies. Curr. Opin. Plant Biol. 2011;14:130–136. doi: 10.1016/j.pbi.2011.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saxena A, Carninci P. Whole transcriptome analysis: what are we still missing? Wiley Interdiscip Rev. Syst. Biol. Med. 2011;3:527–543. doi: 10.1002/wsbm.135. [DOI] [PubMed] [Google Scholar]
- 28.Engelen S, Tahi F. Tfold: efficient in silico prediction of non-coding RNA secondary structures. Nucleic Acids Res. 2010;38:2453–2466. doi: 10.1093/nar/gkp1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gruber AR, Findeiβ S, Washietl S, Hofacker IL, Stadler PF. RNAz 2.0: Improved noncoding RNA detection. Pac. Symp. Biocomput. 2010;15:69–79. [PubMed] [Google Scholar]