Abstract
Only a small fraction of known proteins have been functionally characterized, making protein function prediction essential to propose annotations for uncharacterized proteins. In recent years many function prediction methods have been developed using various sources of biological data from protein sequence and structure to gene expression data. Here we present the CombFunc web server, which makes Gene Ontology (GO)-based protein function predictions. CombFunc incorporates ConFunc, our existing function prediction method, with other approaches for function prediction that use protein sequence, gene expression and protein–protein interaction data. In benchmarking on a set of 1686 proteins CombFunc obtains precision and recall of 0.71 and 0.64 respectively for gene ontology molecular function terms. For biological process GO terms precision of 0.74 and recall of 0.41 is obtained. CombFunc is available at http://www.sbg.bio.ic.ac.uk/combfunc.
INTRODUCTION
Protein function prediction is essential to provide insight to the functions of uncharacterized proteins. This is highlighted by the gap between the large number of proteins that have been identified and the small percentage of them that have been functionally characterized (1). Annotation transfer using BLAST (2) represents a standard and widely-used method of function prediction but as protein function is often only conserved by homologues sharing a high sequence identity, this approach can be prone to errors (3). In recent years many methods have been developed to improve upon BLAST-based annotation transfer. This has included methods such as GOtcha (4) and PFP/ESG (5,6), which combine the Gene Ontology (GO) (7) annotations present in multiple homologues and use their e-values to weight predictions or use machine learning to optimize predictions (8). Phylogenomics approaches distinguish between orthologues and paralogues to infer function (9). The presence of domains from Interpro (10) or Pfam (11) are used for electronic annotation in GO annotations (12) and combinations of domains have also been used for function prediction (13). In ConFunc we used conserved residues representative of individual GO terms to predict protein function (14). Other methods have used protein–protein interaction networks (15,16), gene co-expression (17,18) or multiple protein features including protein disorder and secondary structure (19).
Some methods combine predictions from multiple sources of data (20–26). This includes methods that use Bayesian approaches (24) or Support Vector Machines (SVMs) (23,25) to combine predictions. Some of these methods are available as web servers. The ProKnow (21) webserver combines the evidence from multiple sources to make overall predictions of GO functions. In contrast the ProFunc (20) and PredUS (22) servers do not make overall predictions of protein function, instead they enable the user to explore the results of the many sequence and structural analyses that they perform. Further details of these methods and others are available in recent reviews (1,27).
Here we present CombFunc a server for GO-based protein function prediction. CombFunc incorporates ConFunc (14) our existing sequence based function prediction method and it also extends our recent use of multiple methods to predict the functions of proteins in the Plasmodium berghei male gamete (28). CombFunc uses sequence information including BLAST/PSI-BLAST (29) annotation transfer, domain information from Interpro, protein–protein interaction data from IntAct (30) and MiNT (31) and gene expression data from COXPRESdb (32).
MATERIALS AND METHODS
The CombFunc algorithm
CombFunc obtains information from multiple analyses which are then combined using a SVM (33) to make an overall prediction. The data sources used are described below.
The sequence-based sources of input to CombFunc are: ConFunc, BLAST/PSI-BLAST annotation transfer, domain information and a sequence search against the fold library of Phyre2 (34), our in-house protein structure prediction server. ConFunc is run as previously described in Wass and Sternberg (14). Both BLAST and PSI-BLAST are used to search for GO annotated homologues of the query sequence in UniProt (35). Where PSI-BLAST is used, UniRef50 is initially searched and the profile generated is used to search the full UniProt database as this approach has been shown to improve the identification of homologues (36). Domain information is obtained using Interpro (10) and Pfam domain combinations are also used to make predictions as described in (13). HHsearch (37) is used to search the fold library of Phyre2 to identify structures homologous to the query sequence, whose annotations are input to the SVM. All methods use only experimentally determined GO annotations.
The non-sequence-based data sources are protein–protein interactions (PPI) and gene co-expression. PPI data are obtained from both IntAct (30) and MiNT (31). Function prediction is performed by simple neighbour counting (38) and indirect neighbours are also included (15). Gene expression data is obtained from the COXPRESdb database (32), which contains expression data for Human, Mouse, Rat, Chicken, Zebrafish, Fly and Nematode. COXPRESdb uses a mutual rank score to determine the strength of co-expression, which is calculated as the geometric mean of the correlation rank of gene A to gene B and of gene B to gene A. The frequency of GO terms within the set of co-expressed genes with a mutual rank less than 50 (39) is input into the SVM.
CombFunc uses each of the individual methods to identify GO terms that may be associated with the query. Features associated with the GO terms identified by individual methods are used by CombFunc to make a final prediction of the query function. The features used for each method are listed in Supplementary Table S1 and are described below.
For BLAST and PSI-BLAST the top annotated hit is identified and the GO terms it is annotated with are used for prediction. The features from BLAST and PSI-BLAST include the e-value of the top annotated hit, the sequence identity between the query and top annotated hit and also the sequence coverage of the query by the top hit. Additionally for PSI-BLAST data the annotations of multiple sequences are considered by calculating the i-score as used in GOtcha (4). For terms identified by the interactome analysis the features correspond to the fraction of direct and also indirect neighbours that are annotated with that term. For terms present in the Interpro analysis, the feature corresponds to the lowest e-value of a domain hit annotated with that term (maximum of 1). For the Pfam domain combinations analysis the feature is 1 if predicted by the method and 0 otherwise. Features from the Phyre2 fold library use terms present in the top annotated hit and use the probability score from HHsearch (37) between the query and the hit and also the sequence coverage of the query by the hit. Features for GO terms identified from expression data use a number of features including: the fraction of co-expressed proteins annotated with the function and the minimum, average and maximum mutual rank and correlation coefficients of the co-expressed proteins. Finally a feature is included for each of the individual level 1 GO terms (i.e. binding and catalytic function in molecular function). These features are set to 1 if they are a parent term of the term being considered and zero otherwise.
CombFunc uses three classifiers for the molecular function and biological process categories. As the features associated with GO terms are likely to vary depending on their location in the GO graph, the three classifiers are used for different levels of GO. One classifier considers only terms one level below the root (e.g. catalytic activity or binding in the molecular function category), the second considers terms in the next two levels, while the third classifier considers all more specific terms. The scores output from the SVMs are converted to probabilities as described in Platt (40). The classification process is repeated 10 times, using the 10 sets of optimized SVMs generated during cross-validation. GO terms are predicted to be a function of the query protein if they are predicted to be so by at least 5 of the 10 sets of SVMs with a probability score set as an average of the probability scores for the SVMs that predicted the function.
Generating a test set
A test set of proteins with experimental GO annotations in both the molecular function and biological process GO categories was extracted using the UniProt-GOA annotations from December 2011. This was reduced to a representative set with less than or equal to 25% sequence identity using CD-HIT (41). Of the resulting 6686 sequences, 5000 were used for cross-validation and the remaining 1686 for final testing of the server.
SVM training
The SVMs were generated using SVMlight (33). A linear kernel was used for classification. For each of the 10-fold, eight were used for training, a further fold was used for optimization and the SVM tested on the remaining fold. In cross-validation each SVM was optimized for the trade off between training error and margin. As the training data is unbalanced with many more negative examples than positive ones we also assessed the effect of the cost factor to identify how training errors on positive examples should outweigh those on negative examples (see Supplementary Material section).
EVALUATING COMBFUNC PERFORMANCE
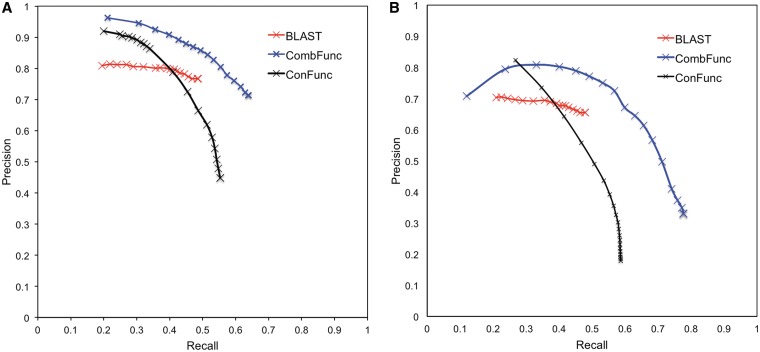
Here we assess the performance of CombFunc using the set of sequences that were not used in cross-validation. The performance of CombFunc on this set of 1686 sequences was assessed using precision and recall calculated as described in Wass and Sternberg (14). The precision-recall graphs in Figure 1 show the performance of CombFunc at a range of thresholds and a comparison with the performance of BLAST annotation transfer. For CombFunc the performance is assessed at confidence thresholds in the range 0–1. We observe that at high confidence (>0.95) CombFunc obtains high precision (0.96) and low recall (0.21). As the threshold is reduced the recall increases while precision reduces and including low confidence predictions CombFunc obtains precision and recall of 0.71 and 0.64 respectively (Figure 1A). CombFunc does not perform as well on biological process terms with both lower recall and precision at equivalent confidence scores. Using a confidence threshold of 0.3 obtains precision of 0.74 and recall of 0.41.
Figure 1.
Benchmarking CombFunc. Precision-recall graphs showing the performance of CombFunc on 1686 sequences not used in cross-validation. CombFunc results are shown in blue, ConFunc in black and BLAST in red. For (A) the GO molecular function and (B) biological process categories.
For comparison the performance of BLAST and ConFunc on the same dataset was considered. For BLAST (Version 2.219) annotation transfer the UniProt database (version December 2011) was searched and the annotation of the top (lowest e-value) experimentally annotated hit transferred to the query sequence. A range of precision and recall scores is obtained by only transferring the annotation if the top hit has an e-value below a threshold, which was varied from 0 − 1e−03. For ConFunc precision-recall values were obtained using a threshold for the ratio score (range 0–1). For benchmarking of all three methods, sequences with >99% sequence identity were excluded for the sequence based prediction components to ensure that the query sequence was not used to make predictions for itself.
We observe that CombFunc performs better than both BLAST and ConFunc. For ConFunc predictions there is a large reduction in precision as the prediction threshold is reduced. ConFunc considers all of the annotations that are present in the homologues of the query identified by BLAST. This often includes the annotation of the query sequence but additionally includes many other functions that are not annotations of the query sequence. At low thresholds many false positive predictions are made. In contrast through the use of multiple data sources and machine learning, CombFunc does not have such a large reduction in precision at lower thresholds, particularly when predicting molecular function terms (Figure 1).
THE COMBFUNC WEB SERVER
CombFunc is available at http://www.sbg.bio.ic.ac.uk/combfunc. Users are required to submit a protein sequence in fasta format and they may also input the UniProt accession of the query sequence. The UniProt accession is required to perform the PPI and co-expression analyses. Processing time for each submission can vary from between 20 min to a few hours, this is largely due to the time taken to perform the search of the Phyre2 fold library.
Results output
CombFunc results output is split into two main sections. The prediction section provides details of the functions predicted by the SVM. In the second section details of the data generated from each of the individual analyses are provided, which users can explore to obtain further details of the data used to make the prediction.
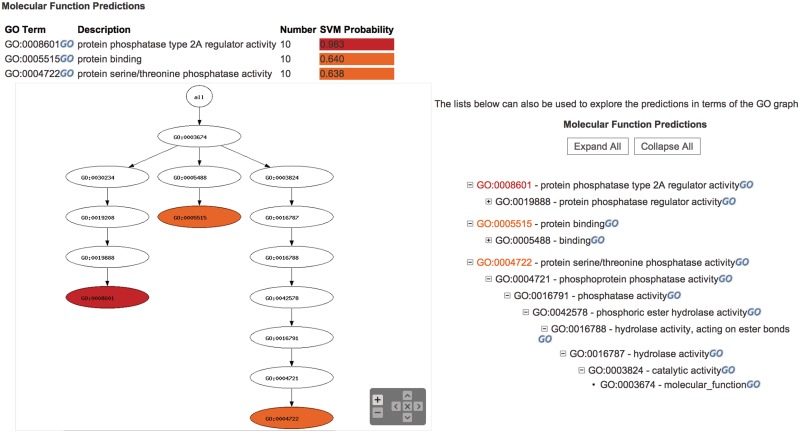
The prediction section displays separate results for molecular function and biological process predictions. For both of these GO categories a table of the predictions lists the term, its name and the probability score of the prediction, this has a range of 0–1, with 1 being the highest confidence (Figure 2). The probability scores are colour coded to indicate the confidence of the predictions, ranging from yellow for low probability predictions to red for high probability. Longer descriptions of the predicted functions are displayed adjacent to the table when the mouse is moved over the rows of the table. Additionally links to the GO terms on the GO website are provided, enabling the user to access external further information about the predicted GO terms.
Figure 2.
Display of a CombFunc prediction. CombFunc predictions are displayed in a table showing the confidence of the prediction and in an image and list placing them in the context of GO structure.
The predictions are visualized within the GO graph in an image that displays a subgraph of GO containing all of the predicted terms and their parent terms (Figure 2). Again predicted terms are colour coded to indicate the confidence of their prediction. The image has a zoom function that enables users to zoom into different areas of the graph to investigate the predictions, which is particularly useful when multiple terms are predicted and the subgraph becomes large. Additionally, the predictions are displayed as an expandable list, which enables similar investigation of the predicted terms.
The second section of the results page contains the output from each of the individual analyses performed. The data associated with each analysis are initially hidden so that the user can view only the analyses they wish to. For each analysis a table lists the GO terms identified by the method and the values or scores associated with those terms. Interpro results are additionally displayed graphically enabling the user to identify the location of the hits on the query sequence. For all analyses the same colour coding as for the main predictions is used to give an indication of how ‘good’ the different scores displayed are. This includes colour coding sequence identity and e-values of BLAST hits and mutual rank values for gene co-expression. Where relevant, links to external data on the GO, UniProt and Intpero websites are provided.
For each submission to CombFunc a submission is also made to 3DLigandSite (42,43), our in-house ligand binding site prediction server. This enables users to combine the function prediction results with the binding site prediction of 3DLigandSite. A link to the 3DligandSite results is provided at the end of the analysis section.
CONCLUSION
CombFunc was developed to utilize the multiple data sources that are available for protein function prediction. In benchmarking CombFunc obtains good performance with 0.71 and 0.64 precision and recall respectively for molecular function GO terms and precision of 0.74 and recall of 0.41 for biological process terms. The CombFunc server provides a resource for users to view predicted functions in both tabular and graphical formats, access to the raw data from each individual method and access to external resources to enable users to explore the functions and data used to make predictions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table 1 and Supplementary Methods.
FUNDING
Biotechnology and Biological Sciences Research Council [BB/F020481/1 to M.N.W.]. Funding for open access charge: Imperial College London Library.
Conflict of interest statement. M.J.E.S. is a founder director of Equinox Pharma Ltd, holds shares in the company, and has obtained remuneration from the company. Equinox Pharma Ltd is exploiting computational methods for drug discovery and markets software.
ACKNOWLEDGEMENTS
The authors would like to thank Lawrence Kelley for advice on the use of SVMs, Suhail Islam for technical support and Michael Tress for helpful discussions about function prediction.
REFERENCES
- 1.Erdin S, Lisewski AM, Lichtarge O. Protein function prediction: towards integration of similarity metrics. Curr. Opin. Struct. Biol. 2011;21:180–188. doi: 10.1016/j.sbi.2011.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 3.Devos D, Valencia A. Practical limits of function prediction. Proteins. 2000;41:98–107. [PubMed] [Google Scholar]
- 4.Martin DM, Berriman M, Barton GJ. GOtcha: a new method for prediction of protein function assessed by the annotation of seven genomes. BMC bioinformatics. 2004;5:178. doi: 10.1186/1471-2105-5-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hawkins T, Chitale M, Luban S, Kihara D. PFP: automated prediction of gene ontology functional annotations with confidence scores using protein sequence data. Proteins. 2009;74:566–582. doi: 10.1002/prot.22172. [DOI] [PubMed] [Google Scholar]
- 6.Chitale M, Hawkins T, Park C, Kihara D. ESG: extended similarity group method for automated protein function prediction. Bioinformatics. 2009;25:1739–1745. doi: 10.1093/bioinformatics/btp309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clark WT, Radivojac P. Analysis of protein function and its prediction from amino acid sequence. Proteins. 2011;79:2086–2096. doi: 10.1002/prot.23029. [DOI] [PubMed] [Google Scholar]
- 9.Engelhardt BE, Jordan MI, Srouji JR, Brenner SE. Genome-scale phylogenetic function annotation of large and diverse protein families. Genome Res. 2011;21:1969–1980. doi: 10.1101/gr.104687.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hunter S, Jones P, Mitchell A, Apweiler R, Attwood TK, Bateman A, Bernard T, Binns D, Bork P, Burge S, et al. InterPro in 2011: new developments in the family and domain prediction database. Nucleic Acids Res. 2012;40:D306–D312. doi: 10.1093/nar/gkr948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, Pang N, Forslund K, Ceric G, Clements J, et al. The Pfam protein families database. Nucleic Acids Res. 2012;40:D290–D301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dimmer EC, Huntley RP, Alam-Faruque Y, Sawford T, O'Donovan C, Martin MJ, Bely B, Browne P, Mun Chan W, Eberhardt R, et al. The UniProt-GO annotation database in 2011. Nucleic Acids Res. 2012;40:D565–D570. doi: 10.1093/nar/gkr1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Forslund K, Sonnhammer EL. Predicting protein function from doma in content. Bioinformatics. 2008;24:1681–1687. doi: 10.1093/bioinformatics/btn312. [DOI] [PubMed] [Google Scholar]
- 14.Wass MN, Sternberg MJ. ConFunc–functional annotation in the twilight zone. Bioinformatics. 2008;24:798–806. doi: 10.1093/bioinformatics/btn037. [DOI] [PubMed] [Google Scholar]
- 15.Chua HN, Sung WK, Wong L. Exploiting indirect neighbours and topological weight to predict protein function from protein-protein interactions. Bioinformatics. 2006;22:1623–1630. doi: 10.1093/bioinformatics/btl145. [DOI] [PubMed] [Google Scholar]
- 16.Vazquez A, Flammini A, Maritan A, Vespignani A. Global protein function prediction from protein-protein interaction networks. Nat. Biotechnol. 2003;21:697–700. doi: 10.1038/nbt825. [DOI] [PubMed] [Google Scholar]
- 17.Brown MP, Grundy WN, Lin D, Cristianini N, Sugnet CW, Furey TS, Ares M, Jr, Haussler D. Knowledge-based analysis of microarray gene expression data by using support vector machines. Proc. Natl Acad. Sci. USA. 2000;97:262–267. doi: 10.1073/pnas.97.1.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl Acad. Sci. USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lobley A, Swindells MB, Orengo CA, Jones DT. Inferring function using patterns of native disorder in proteins. PLoS Computat. Biol. 2007;3:e162. doi: 10.1371/journal.pcbi.0030162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laskowski RA, Watson JD, Thornton JM. ProFunc: a server for predicting protein function from 3D structure. Nucleic Acids Res. 2005;33:W89–W93. doi: 10.1093/nar/gki414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pal D, Eisenberg D. Inference of protein function from protein structure. Structure London. 2005;13:121–130. doi: 10.1016/j.str.2004.10.015. [DOI] [PubMed] [Google Scholar]
- 22.Zhang QC, Deng L, Fisher M, Guan J, Honig B, Petrey D. PredUs: a web server for predicting protein interfaces using structural neighbors. Nucleic Acids Res. 2011;39:W283–W287. doi: 10.1093/nar/gkr311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guan Y, Myers CL, Hess DC, Barutcuoglu Z, Caudy AA, Troyanskaya OG. Predicting gene function in a hierarchical context with an ensemble of classifiers. Genome Biol. 2008;9(Suppl. 1):S3. doi: 10.1186/gb-2008-9-s1-s3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Troyanskaya OG, Dolinski K, Owen AB, Altman RB, Botstein D. A Bayesian framework for combining heterogeneous data sources for gene function prediction (in Saccharomyces cerevisiae) Proc. Natl Acad. Sci. USA. 2003;100:8348–8353. doi: 10.1073/pnas.0832373100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Obozinski G, Lanckriet G, Grant C, Jordan MI, Noble WS. Consistent probabilistic outputs for protein function prediction. Genome Biol. 2008;9(Suppl. 1):S6. doi: 10.1186/gb-2008-9-s1-s6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nariai N, Kolaczyk ED, Kasif S. Probabilistic protein function prediction from heterogeneous genome-wide data. PloS One. 2007;2:e337. doi: 10.1371/journal.pone.0000337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gherardini PF, Helmer-Citterich M. Structure-based function prediction: approaches and applications. Brief. Funct. Genom Proteomics. 2008;7:291–302. doi: 10.1093/bfgp/eln030. [DOI] [PubMed] [Google Scholar]
- 28.Wass MN, Stanway R, Blagborough AM, Lal K, Prieto JH, Raine D, Sternberg MJ, Talman AM, Tomley F, Yates J, et al. Proteomic analysis of Plasmodium in the mosquito: progress and pitfalls. Parasitology. 2012 doi: 10.1017/S0031182012000133. February 16 (doi:10.1017/S0031182012000133; epub ahead of print). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kerrien S, Aranda B, Breuza L, Bridge A, Broackes-Carter F, Chen C, Duesbury M, Dumousseau M, Feuermann M, Hinz U, et al. The IntAct molecular interaction database in 2012. Nucleic Acids Res. 2012;40:D841–D846. doi: 10.1093/nar/gkr1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Licata L, Briganti L, Peluso D, Perfetto L, Iannuccelli M, Galeota E, Sacco F, Palma A, Nardozza AP, Santonico E, et al. MINT, the molecular interaction database: 2012 update. Nucleic Acids Res. 2012;40:D857–D861. doi: 10.1093/nar/gkr930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Obayashi T, Kinoshita K. COXPRESdb: a database to compare gene coexpression in seven model animals. Nucleic Acids Res. 2011;39:D1016–D1022. doi: 10.1093/nar/gkq1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vapnik VN. An overview of statistical learning theory. IEEE Transact Neural Networks / a publication of the IEEE Neural Networks Council. 1999;10:988–999. doi: 10.1109/72.788640. [DOI] [PubMed] [Google Scholar]
- 34.Kelley LA, Sternberg MJ. Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protocols. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 35.Ongoing and future developments at the Universal Protein Resource. Nucleic Acids Res. 39:D214–D219. doi: 10.1093/nar/gkq1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chubb D, Jefferys BR, Sternberg MJ, Kelley LA. Sequencing delivers diminishing returns for homology detection: implications for mapping the protein universe. Bioinformatics. 2010;26:2664–2671. doi: 10.1093/bioinformatics/btq527. [DOI] [PubMed] [Google Scholar]
- 37.Soding J. Protein homology detection by HMM-HMM comparison. Bioinformatics. 2005;21:951–960. doi: 10.1093/bioinformatics/bti125. [DOI] [PubMed] [Google Scholar]
- 38.Schwikowski B, Uetz P, Fields S. A network of protein-protein interactions in yeast. Nat. Biotechnol. 2000;18:1257–1261. doi: 10.1038/82360. [DOI] [PubMed] [Google Scholar]
- 39.Obayashi T, Hayashi S, Shibaoka M, Saeki M, Ohta H, Kinoshita K. COXPRESdb: a database of coexpressed gene networks in mammals. Nucleic Acids Res. 2008;36:D77–82. doi: 10.1093/nar/gkm840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Platt JC. Advances in Large Margin Classifiers. Vol. 1. Cambridge, Massachusetts, US: MIT Press; 1999. pp. 61–74. [Google Scholar]
- 41.Huang Y, Niu B, Gao Y, Fu L, Li W. CD-HIT Suite: a web server for clustering and comparing biological sequences. Bioinformatics. 2010;26:680–682. doi: 10.1093/bioinformatics/btq003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wass MN, Kelley LA, Sternberg MJ. 3DLigandSite: predicting ligand-binding sites using similar structures. Nucleic Acids Res. 2010;38:W469–W473. doi: 10.1093/nar/gkq406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wass MN, Sternberg MJ. Prediction of ligand binding sites using homologous structures and conservation at CASP8. Proteins. 2009;77(Suppl. 9):147–151. doi: 10.1002/prot.22513. [DOI] [PMC free article] [PubMed] [Google Scholar]