Summary
Migration of fragmented mitochondrial DNA (mtDNA) to the nucleus has been shown to occur in multiple species including yeast, plants, and mammals. Several human diseases, including Pallister-Hall syndrome and mucolipidosis, can be initiated by mtDNA insertion mutagenesis of nuclear DNA. In yeast, we demonstrated that the rate of mtDNA fragments translocating to the nucleus increases during chronological aging. The yeast chronological life span (CLS) is determined by the survival of non-dividing cell populations. Whereas yeast strains with elevated migration rates of mtDNA fragments to the nucleus showed accelerated chronological aging, strains with decreased mtDNA transfer rates to the nucleus exhibited an extended CLS. Although one of the most popular theories of aging is the free radical theory, migration of mtDNA fragments to the nucleus may also contribute to the chronological aging process by possibly increasing nuclear genomic instability in cells with advanced age.
Keywords: aging, chromosomal DNA, chronological aging, chronological life span, DNA damage, mitochondrial DNA, mtDNA, yeast
Reactive oxygen species (ROS) may escape from the mitochondrial respiration process and contribute to an increase in oxidative stress leading to accelerated aging (Schriner et al., 2005). Here, we hypothesize that the translocation of fragmented mitochondrial DNA (mtDNA) to the nucleus also contributes to the aging process.
Mitochondria were once free-living prokaryotes. Many genes have relocated from the ancestral organellar genomes to the nucleus during the two billion years since eukaryotes arose (Leister, 2005). Most (>98%) mitochondrial proteins are now encoded by nuclear DNA, synthesized on cytoplasmic ribosomes and then imported into mitochondria. MtDNA encodes only few proteins (many of those are members of the respiratory chain), some transfer RNAs and ribosomal RNAs (Anderson et al., 1981; Foury et al., 1998). The presence of sequence elements encoding mtDNA at numerous sites in a variety of eukaryotic nuclear genomes (called nuclear DNA sequences of mitochondrial origin or NUMTs) are further indications for the evolutionary transfer of mtDNA fragments to the nucleus (Gherman et al., 2007; Hazkani-Covo et al., 2010; Ricchetti et al., 2004).
However, recent evidence has suggested that the transfer of mtDNA to the nucleus is still an ongoing process leading to de novo disruptions of nuclear genes (Hazkani-Covo et al., 2010; Ricchetti et al., 2004; Shay and Werbin, 1992; Turner et al., 2003). Insertions of mtDNA fragments into human chromosomal DNA do or may contribute to the development of several diseases including Pallister-Hall syndrome and mucolipidosis (Borensztajn et al., 2002; Goldin et al., 2004; Turner et al., 2003; Willett-Brozick et al., 2001). Experiments with yeast have demonstrated that fragmented mtDNA is captured during the repair of induced double-stranded DNA breaks in nuclear chromosomal or plasmid DNA (Decottignies, 2005; Ricchetti et al., 1999; Yu and Gabriel, 1999).
Using Saccharomyces cerevisiae (baker’s yeast) as a model system, we investigated whether the frequency of mtDNA fragments migrating to the nucleus alters during the chronological aging process. The chronological life span (CLS) in yeast is defined as the length of time cells survive after they are driven into a non-dividing state (G0-arrest) by depletion of nutrients from the growth medium (Bitterman et al., 2003; Fabrizio and Longo, 2003). The non-dividing state of nutrient-depleted yeast cells shares features with post-mitotic differentiated cells in higher eukaryotes. Chronologically aging yeast is a simple system to investigate how somatic cells are maintained and how spontaneous mutations are induced in non-dividing cells.
We determined the transfer rate of mtDNA fragments to the nucleus by a genetic assay using a strain that was constructed in Dr. Tom Fox’s laboratory (Cornell U of Ithaca, NY) (Thorsness and Fox, 1993). This strain contains the nuclear yeast TRP1 gene inserted into mtDNA (Figure 1A). The endogenous nuclear TRP1 gene is deleted in this strain. The mitochondrial TRP1 gene becomes only functional after its transfer to the nucleus, because the required transcription machinery is present in the nucleus, but not in mitochondria. The cells, in which the mitochondrial TRP1 gene has moved to the nucleus, can be selected on growth medium lacking the amino acid tryptophan (Supplementary Figure 1, bottom row). In contrast, the control strain without the mitochondrial TRP1 gene does not grow in the absence of tryptophan.
Figure 1. The migration frequency of mtDNA fragments to the nucleus increases during the yeast chronological life span.
(A) Scheme explaining the assay for the measurement of the mtDNA transfer frequency to the nucleus in yeast.
(B) Yeast cells were grown in rich, glucose-containing YEPD medium. To determine the total number of viable cells in the aging culture (CLS), appropriate dilutions of cells were spread on YEPD plates at the indicated days (left panel). To determine the mtDNA transfer frequency (right panel), 5x107 cells were plated on plates lacking tryptophan on the same days. The numbers of colonies obtained from those plates were then normalized to the numbers of viable cells. The average migration frequency of mtDNA to the nucleus increases from 2.6 (day 3) to 77 (day 29) events per 106 viable cells. The control strain lacked the mitochondrial TRP1 gene. Standard errors of three independent experiments are shown.
(C) Yeast cells were grown in defined, synthetic glucose-containing medium (YC). The CLS and the mtDNA transfer frequency were determined as explained in panel B. The average migration frequency of mtDNA to the nucleus increases from 2.5 (day 3) to 15.3 (day 14) events per 106 viable cells. The control strain lacked the mitochondrial TRP1 gene. Standard errors of three independent experiments are shown.
The survival rates (CLS) were determined by spreading out appropriate numbers of cells on plates containing rich growth medium (Supplementary Figure 1, top row). The mitochondrial TRP1-containing strain and the control strain showed similar CLS (Figure 1B, left panel), indicating that the presence of the nuclear TRP1 marker gene in mtDNA does not influence the CLS. To determine the mtDNA transfer rates on the same days, equal numbers of cells were spread on plates lacking tryptophan (Supplementary Figure 1, bottom row). The numbers of colonies that could grow on the plates lacking tryptophan were normalized to the total living cell count (survival rate). We observed an about 30-fold increase of the mtDNA transfer rate during the CLS (day 3 compared to day 29; Figure 1B, right panel). These results suggest that during the CLS the number of mtDNA fragments migrating to the nucleus increases. Whereas in Figure 1B the CLS of cells grown in rich medium (YEPD) are shown, similar trends were obtained with the same strains grown in synthetic growth medium (YC; Figure 1C). We also analyzed the nuclear mtDNA-TRP1 molecules of individual transfer events by Southern technique (Supplementary Figure 2). The sizes of the DNA molecules decreased in the terminal phase of the CLS suggesting that increased mtDNA fragmentation occurs in later phases of the CLS (Supplementary Figure 2, graph). Further we used the two-dimensional agarose gel electrophoresis technique to demonstrate that the nuclear mtDNA-TRP1 molecules are primarily present as circular molecules as previously suggested (Supplementary Figure 3) (Ivessa et al., 2000; Thorsness and Fox, 1993). Although the nuclear mtDNA-TRP1 molecules appear to exist mostly as extrachromosomal circles throughout the CLS, we can demonstrate by using the pulse-field-gel-electrophoresis technique that a fraction of the mtDNA fragments that enter the nucleus integrate also into chromosomal DNA (Supplementary Figure 4). The frequency of the insertion events of mtDNA fragments into nuclear DNA is elevated in later phases of the CLS.
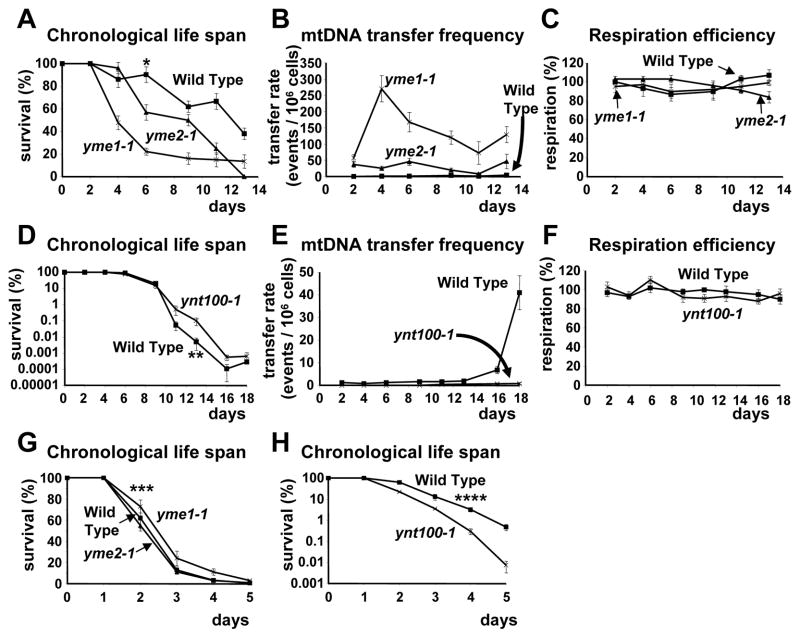
To determine whether the observed increase in the migration rate of mtDNA fragments to the nucleus during chronological aging has a direct effect on aging, we used mutant yeast strains with known increased or decreased mtDNA translocation rates to the nucleus. Whereas the yme (yeast mtDNA escape) mutants exhibit elevated translocation frequencies, the ynt100-1 mutant shows a lower migration frequency compared to wild type (WT) cells (Shafer et al., 1999; Thorsness and Fox, 1993). The yme and ynt100-1 mutant strains are all respiratory efficient and can grow on glycerol-containing growth medium at 30°C (Shafer et al., 1999; Thorsness and Fox, 1993). We used the yme1-1 and yme2-1 mutant strains in our analyses. Both Yme1p, which is a protease, and Yme2p are mitochondrial proteins (Hanekamp and Thorsness, 1996; Thorsness et al., 1993). The identity of the ynt100-1 mutant is currently unknown. In Figure 2, we demonstrated that the yme1-1 and yme2-1 mutant strains have an about 50% and 25%, respectively, shorter CLS, and an average 77-fold and 17–fold, respectively, higher translocation rate of mtDNA fragments to the nucleus compared to WT. As previously reported the yme1-1 mutation causes also shortening of the CLS in the BY4741 strain background (Palermo et al., 2007). In contrast, the ynt100-1 mutant strain, which has a lower mtDNA translocation rate to the nucleus, exhibited a slightly extended CLS in the terminal phase (Figure 2, panel D). These results suggest that the increased mtDNA translocation rate to the nucleus, as observed in Figure 1, promotes aging. All mutant strains had respiration efficiencies throughout the CLS that were comparable to WT (Figure 2, panels C and F).
Figure 2. The translocation of mtDNA fragments to the nucleus affects the yeast chronological life span.
The strains were grown in defined, synthetic glucose-containing medium. To determine the total numbers of viable cells during the CLS, appropriate dilutions of cells were spread on plates containing rich medium (YEPD) at the indicated days (panels A, D, G, and H). To determine the mtDNA transfer frequency, 5x107 cells were plated on plates lacking tryptophan on the same days (panels B and E). The numbers of colonies obtained from those plates were then normalized to the numbers of viable cells. The respiration efficiency is expressed as a ratio of the number of colonies obtained from glycerol-containing plates (respiring cells) to the number of colonies obtained from glucose-containing plates (total viable cells) (panels C and F). The strains shown in panels A to F contain functional mtDNA (rho+), whereas the strains shown in panels G and H lack mtDNA (rho0). Standard errors of three independent experiments are shown. The significance in the difference in the CLS was determined by a Student’s t-test (P < 0.005 (*) for day 6 in panel A, P < 0.01 (**) for day 13 in panel D, P < 0.4 (***) for day 2 in panel G, and P < 0.01 (****) for day 4 in panel H).
To determine whether the observed aging phenotypes are indeed dependent on mtDNA we used WT and mutant strains lacking mtDNA (rho0). Whereas the yme1-1 and yme2-1 rho0 mutant strains had similar short CLS as a WT rho0 strain, the ynt100-1 rho0 mutant strain had an even slightly shorter CLS compared to the WT rho0 strain (Figures 2G and H). These results suggest that the shortening or extension of the CLS as observed in the yme and the ynt100-1 strains, respectively, is dependent to a large part on the presence of mtDNA. Consistent with our results others also demonstrated that depletion of mtDNA shortens the CLS and explained this phenotype that yeast cells lacking mtDNA have difficulties in the switch from fermentation to respiration during the diauxic shift (Buttner et al., 2008). Mitochondrial dysfunction decreases the CLS as it has been reported for both baker’s and fission yeast (Aerts et al., 2009; Zuin et al., 2008). Mice expressing a proof-reading domain defective mtDNA polymerase γ gene exhibit also a decreased life span (Kujoth et al., 2005; Trifunovic et al., 2004). Although these reports suggest that mitochondrial dysfunction and loss of mtDNA lead to a shortening of the lifespan, there are also reports that WT yeast cells lacking mtDNA exhibit a longer CLS compared to cells containing mtDNA (Mazzoni et al., 2005). The authors explained the lengthening of the CLS that the lowering of the cellular concentration of ROS as a result of the lack of mtDNA results in lower oxidative stress levels and a longer CLS. In fact, rho0 strains are resistant to ROS-generating agents and have lower amounts of ROS (Davermann et al., 2002; Rasmussen et al., 2003).
Future studies will determine whether the insertions of mtDNA fragments into nuclear DNA increase nuclear genomic instability and promote aging. However, the extrachromosomal mtDNA circles in the nucleus may also contribute to the aging process by, for example, binding nuclear proteins which are normally required for maintaining the stability of chromosomal DNA. Since the nuclear mutation rate including base substitutions, frame shift mutations, and gross-chromosomal rearrangements increase during chronological aging, there is the possibility that mtDNA fragments migrating to the nucleus also contribute to this increase (Madia et al., 2008).
Supplementary Material
Acknowledgments
We are grateful to Drs. Tom Fox (Cornell U of Ithaca, NY) and Peter Thorsness (U of Wyoming, Laramie, WY) for providing strains, to Dr. David Kaback for letting us use his PFGE equipment, and to Dr. Edouard Azzam for critical reading of the manuscript. This work is supported by a Scientist Development Grant from the American Heart Association (0735174N) and in part by a Program Project Grant from the National Institute of Health (5P01AG027211).
References
- Aerts AM, Zabrocki P, Govaert G, Mathys J, Carmona-Gutierrez D, Madeo F, Winderickx J, Cammue BP, Thevissen K. Mitochondrial dysfunction leads to reduced chronological lifespan and increased apoptosis in yeast. FEBS Lett. 2009;583:113–117. doi: 10.1016/j.febslet.2008.11.028. [DOI] [PubMed] [Google Scholar]
- Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, et al. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- Bitterman KJ, Medvedik O, Sinclair DA. Longevity regulation in Saccharomyces cerevisiae: linking metabolism, genome stability, and heterochromatin. Microbiol Mol Biol Rev. 2003;67:376–399. doi: 10.1128/MMBR.67.3.376-399.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borensztajn K, Chafa O, Alhenc-Gelas M, Salha S, Reghis A, Fischer AM, Tapon-Bretaudiere J. Characterization of two novel splice site mutations in human factor VII gene causing severe plasma factor VII deficiency and bleeding diathesis. Br J Haematol. 2002;117:168–171. doi: 10.1046/j.1365-2141.2002.03397.x. [DOI] [PubMed] [Google Scholar]
- Buttner S, Bitto A, Ring J, Augsten M, Zabrocki P, Eisenberg T, Jungwirth H, Hutter S, Carmona-Gutierrez D, Kroemer G, et al. Functional mitochondria are required for alpha-synuclein toxicity in aging yeast. J Biol Chem. 2008;283:7554–7560. doi: 10.1074/jbc.M708477200. [DOI] [PubMed] [Google Scholar]
- Davermann D, Martinez M, McKoy J, Patel N, Averbeck D, Moore CW. Impaired mitochondrial function protects against free radical-mediated cell death. Free Radic Biol Med. 2002;33:1209–1220. doi: 10.1016/s0891-5849(02)00984-x. [DOI] [PubMed] [Google Scholar]
- Decottignies A. Capture of extranuclear DNA at fission yeast double-strand breaks. Genetics. 2005;171:1535–1548. doi: 10.1534/genetics.105.046144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrizio P, Longo VD. The chronological life span of Saccharomyces cerevisiae. Aging Cell. 2003;2:73–81. doi: 10.1046/j.1474-9728.2003.00033.x. [DOI] [PubMed] [Google Scholar]
- Foury F, Roganti T, Lecrenier N, Purnelle B. The complete sequence of the mitochondrial genome of Saccharomyces cerevisiae. FEBS Lett. 1998;440:325–331. doi: 10.1016/s0014-5793(98)01467-7. [DOI] [PubMed] [Google Scholar]
- Gherman A, Chen PE, Teslovich TM, Stankiewicz P, Withers M, Kashuk CS, Chakravarti A, Lupski JR, Cutler DJ, Katsanis N. Population bottlenecks as a potential major shaping force of human genome architecture. PLoS Genet. 2007;3:e119. doi: 10.1371/journal.pgen.0030119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldin E, Stahl S, Cooney AM, Kaneski CR, Gupta S, Brady RO, Ellis JR, Schiffmann R. Transfer of a mitochondrial DNA fragment to MCOLN1 causes an inherited case of mucolipidosis IV. Hum Mutat. 2004;24:460–465. doi: 10.1002/humu.20094. [DOI] [PubMed] [Google Scholar]
- Hanekamp T, Thorsness PE. Inactivation of YME2/RNA12, which encodes an integral inner mitochondrial membrane protein, causes increased escape of DNA from mitochondria to the nucleus in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:2764–2771. doi: 10.1128/mcb.16.6.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazkani-Covo E, Zeller RM, Martin W. Molecular poltergeists: mitochondrial DNA copies (numts) in sequenced nuclear genomes. PLoS Genet. 2010;6:e1000834. doi: 10.1371/journal.pgen.1000834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivessa AS, Zhou JQ, Zakian VA. The Saccharomyces Pif1p DNA helicase and the highly related Rrm3p have opposite effects on replication fork progression in ribosomal DNA. Cell. 2000;100:479–489. doi: 10.1016/s0092-8674(00)80683-2. [DOI] [PubMed] [Google Scholar]
- Kujoth GC, Hiona A, Pugh TD, Someya S, Panzer K, Wohlgemuth SE, Hofer T, Seo AY, Sullivan R, Jobling WA, et al. Mitochondrial DNA mutations, oxidative stress, and apoptosis in mammalian aging. Science. 2005;309:481–484. doi: 10.1126/science.1112125. [DOI] [PubMed] [Google Scholar]
- Leister D. Origin, evolution and genetic effects of nuclear insertions of organelle DNA. Trends Genet. 2005;21:655–663. doi: 10.1016/j.tig.2005.09.004. [DOI] [PubMed] [Google Scholar]
- Madia F, Gattazzo C, Wei M, Fabrizio P, Burhans WC, Weinberger M, Galbani A, Smith JR, Nguyen C, Huey S, et al. Longevity mutation in SCH9 prevents recombination errors and premature genomic instability in a Werner/Bloom model system. J Cell Biol. 2008;180:67–81. doi: 10.1083/jcb.200707154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzoni C, Herker E, Palermo V, Jungwirth H, Eisenberg T, Madeo F, Falcone C. Yeast caspase 1 links messenger RNA stability to apoptosis in yeast. EMBO Rep. 2005;6:1076–1081. doi: 10.1038/sj.embor.7400514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palermo V, Falcone C, Mazzoni C. Apoptosis and aging in mitochondrial morphology mutants of S. cerevisiae. Folia Microbiol (Praha) 2007;52:479–483. doi: 10.1007/BF02932107. [DOI] [PubMed] [Google Scholar]
- Rasmussen AK, Chatterjee A, Rasmussen LJ, Singh KK. Mitochondria-mediated nuclear mutator phenotype in Saccharomyces cerevisiae. Nucleic Acids Res. 2003;31:3909–3917. doi: 10.1093/nar/gkg446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricchetti M, Fairhead C, Dujon B. Mitochondrial DNA repairs double-strand breaks in yeast chromosomes. Nature. 1999;402:96–100. doi: 10.1038/47076. [DOI] [PubMed] [Google Scholar]
- Ricchetti M, Tekaia F, Dujon B. Continued colonization of the human genome by mitochondrial DNA. PLoS Biol. 2004;2:E273. doi: 10.1371/journal.pbio.0020273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schriner SE, Linford NJ, Martin GM, Treuting P, Ogburn CE, Emond M, Coskun PE, Ladiges W, Wolf N, Van Remmen H, et al. Extension of murine life span by overexpression of catalase targeted to mitochondria. Science. 2005;308:1909–1911. doi: 10.1126/science.1106653. [DOI] [PubMed] [Google Scholar]
- Shafer KS, Hanekamp T, White KH, Thorsness PE. Mechanisms of mitochondrial DNA escape to the nucleus in the yeast Saccharomyces cerevisiae. Curr Genet. 1999;36:183–194. doi: 10.1007/s002940050489. [DOI] [PubMed] [Google Scholar]
- Shay JW, Werbin H. New evidence for the insertion of mitochondrial DNA into the human genome: significance for cancer and aging. Mutat Res. 1992;275:227–235. doi: 10.1016/0921-8734(92)90026-l. [DOI] [PubMed] [Google Scholar]
- Thorsness PE, Fox TD. Nuclear mutations in Saccharomyces cerevisiae that affect the escape of DNA from mitochondria to the nucleus. Genetics. 1993;134:21–28. doi: 10.1093/genetics/134.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorsness PE, White KH, Fox TD. Inactivation of YME1, a member of the ftsH-SEC18-PAS1-CDC48 family of putative ATPase-encoding genes, causes increased escape of DNA from mitochondria in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:5418–5426. doi: 10.1128/mcb.13.9.5418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trifunovic A, Wredenberg A, Falkenberg M, Spelbrink JN, Rovio AT, Bruder CE, Bohlooly YM, Gidlof S, Oldfors A, Wibom R, et al. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature. 2004;429:417–423. doi: 10.1038/nature02517. [DOI] [PubMed] [Google Scholar]
- Turner C, Killoran C, Thomas NS, Rosenberg M, Chuzhanova NA, Johnston J, Kemel Y, Cooper DN, Biesecker LG. Human genetic disease caused by de novo mitochondrial-nuclear DNA transfer. Hum Genet. 2003;112:303–309. doi: 10.1007/s00439-002-0892-2. [DOI] [PubMed] [Google Scholar]
- Willett-Brozick JE, Savul SA, Richey LE, Baysal BE. Germ line insertion of mtDNA at the breakpoint junction of a reciprocal constitutional translocation. Hum Genet. 2001;109:216–223. doi: 10.1007/s004390100564. [DOI] [PubMed] [Google Scholar]
- Yu X, Gabriel A. Patching broken chromosomes with extranuclear cellular DNA. Mol Cell. 1999;4:873–881. doi: 10.1016/s1097-2765(00)80397-4. [DOI] [PubMed] [Google Scholar]
- Zuin A, Gabrielli N, Calvo IA, Garcia-Santamarina S, Hoe KL, Kim DU, Park HO, Hayles J, Ayte J, Hidalgo E. Mitochondrial dysfunction increases oxidative stress and decreases chronological life span in fission yeast. PLoS One. 2008;3:e2842. doi: 10.1371/journal.pone.0002842. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.