Abstract
The G protein-coupled delta opioid receptor gene (dor) has been associated with neuronal survival, differentiation, and neuroprotection. Our previous study identified PI3K/Akt/NF-κB signaling is a main downstream signaling pathway in nerve growth factor (NGF)-induced temporal expression of the dor gene in the PC12 cell model. It is still unknown how NGF/PI3K signaling regulates the expression of the dor gene in the nucleus. In the current study, we investigated how PI3K signaling affected epigenetic modifications of histone H3 Lys9(H3K9) in the 5′-UTR region of the rat dor gene locus. NGF treatment resulted in the global reversal of H3K9 trimethylation in cells. Moreover, the locus-specific reversal of H3K9 trimethylation and acetylation of H3K9 were dependent upon NGF/PI3K signaling and temporally well correlated with NGF-induced gene expression. These results indicate the importance of epigenetic modifications of H3K9, particularly the reversal of trimethylated H3K9, in the regulation of NGF/PI3K-dependent genes during neuronal differentiation.
Keywords: Akt, Chromatin remodeling, Delta opioid receptor, Histone modification, Lysine trimethylation, NF-κB; NGF, PI3K
The G protein-coupled delta opioid receptor (DOR) not only plays a critical role in opioid-mediated pain control, drug tolerance and addiction [1], but it is also associated with neuronal differentiation, cell survival, neuroprotection, and neurogenesis [2-6]. Nerve growth factor (NGF), which is the prototype of neurotrophins, plays a critical role in the development and maintenance of the central and peripheral nervous systems [7]. The rat adrenal pheochromocytoma PC12 cell line is a well-established NGF-responsive model for the study of gene expression during neuronal differentiation and development [8]. NGF temporally induces the expression of the dor gene in PC12 cells[9-11]. Neurotrophins regulate the expression of many neuronal genes during neuronal development. Moreover, the dor gene is expressed both spatially and temporally in specific developmental stages [12,13]. Token together, elucidating the molecular mechanisms of NGF-induced dor gene expression might shed light on the general mechanism of neurotrophin-mediated gene expression during neuronal differentiation and development.
The signal transduction pathways from the cell surface to the nucleus in NGF-responsive PC12 cells have been investigated extensively [14], but the temporal mechanism of NGF-induced gene expression is not well understood at the chromatin level. Phosphorylation and acetylation of histone H3 at the N-terminal position are associated with mitogen-mediated induction of the immediate-early (IE) genes, c-fos and c-jun [15-17]. NGF induces many IE genes and late phase genes in PC12 cells [18]. NGF signaling apparently induces immediate-early genes by the recruitment of transcriptional coactivators/acetyltransferases to chromatin-remodeling complexes on the promoters of induced genes [19]. However, little is known of how NGF induces late phase genes such as dor. We previously observed that sustained activation of phosphatidylinositol 3-kinase (PI3K)/Akt signaling is important for the expression of the late-phase dor gene [11]. It is still unknown how NGF induces the expression of the dor gene after PI3K survival signaling reaches the nucleus.
One possible mechanism for NGF-induced gene expression in the nucleus is that the NGF signal leads to the regulation of locus-specific modifications of histone tails, which subsequently bind to the promoter of an NGF-induced gene to initiate transcription and elongation. Such epigenetic modifications of histone tails can be a critical step for NGF-induced late phase gene expression. To test this idea, we examined the global and locus-specific effects of NGF signaling on modifications of histone H3. We also analyzed the status of H3 trimethylation in the rat dor promoter region in living cells using chromatin immunoprecipitation (ChIP) assays. We provide several lines of evidence showing that NGF treatment results in modifications of H3 Lys9 (H3K9), which correlate temporally with NGF-induced dor gene expression in PC12 cells. NGF/PI3K-mediated gene expression has been associated with a plethora of important biological processes, including cell survival, development, differentiation, and cell cycles [7,20-22]. Hence, NGF/PI3K-induced H3K9 modifications might have broad implications for cell survival, neuronal differentiation and development.
Materials and methods
Cell culture and reagents
The PC12h cell line was a generous gift from Dr. Hiroshi Hatanaka. PC12h cells were grown and maintained as described previously [11]. Nerve Growth Factor (2.5S) was purchased from Harlan Bioproducts for Science, Inc (Indianapolis, IN). LY294002, PD98059, and K-252a were purchased from Calbiochem (La Jolla, CA). Antibodies against H3K9 (H3K9me3), dimethylated H3K9 (H3K9me2), and acetylated H3K9 (H3K9ac) were purchased from Upstate (Lake Placid, NY). Anti-H3 (total) was from Cell Signaling (Beverly, MA).
Western blot analysis
The proteins from cell lysates were separated on either 10% SDS–PAGE gels or 4–20% T Novex® Tris-glycine gels from Invitrogen (Carlsbad, CA) and blotted onto nitrocellular membranes according to the manufacturer’s protocols (Cell Signaling). Post-translationally modified and unmodified protein bands were then probed by the respective antibodies according to the manufacturer’s protocols. Identification and quantification of protein bands were described previously [23] unless otherwise noted. Western blots were processed and figure compositions were constructed using Photoshop®.
Chromatin immunoprecipitation (ChIP)-PCR assays
ChIP assays were performed using the ChIP assay kit according to the manufacture’s protocol (Upstate, Lake Placid, NY). In brief, chromatin was cross-linked with formaldehyde, cells were washed with cold PBS buffer, and the cells were then lysed. DNA was sheared by sonication (three times, 15s each, using the Sonicator™ Model W-220I, Heat Systems Ultrasonics). Cell supernatant was pre-cleared by incubation with G-Sepharose saturated with salmon sperm DNA. The pre-cleared supernatant was incubated with specific primary antibodies overnight. DNA/protein/antibody complex was isolated and analyzed by both semi-quantitative (HotStartTag® PCR from Qiagen, 32–40 cycles) and quantitative real-time PCR (iQ™ SYBR® Green Supermix from Bio-Rad). The primers for the rat dor promoter region around the start codon (ATG), which was identified by pairing the mouse dor promoter sequence [24] and rat dor mRNA sequence (NM_012617) with the rat genomic DNA sequence (NW_047724), are 5′-CTA GAG TCA GCA AAG TCC AGG CAG-3′, 5′-GAC GAT TCC AAA CAT GAC GAG CAC-3′. Quantitative real-time PCR were carried out in triplicate or more to determine the association of acetylated or trimethylated H3K9 with the dor gene. Input DNA was diluted in a fourfold-dilution series to determine the correlation coefficient and PCR efficiency using the Bio-Rad iCycler iQ™ detection system. The fold change in DNA binding to the modified histone H3 relative to input DNA was determined by the equation: Fold change = 2−Δ(ΔCT) where ΔCT = CT(chip) − CT(input) and Δ(ΔCT) = ΔCT(treated) − ΔCT(untreated) or by the individual standard dilution curve for each experiment.
Statistical analysis
When more than two data points were compared, one-way ANOVA was performed for statistical significance evaluation using the GraphPad Prism™ program. Either Dunnett’s or Student’s test was carried out to determine whether or not the sample was significantly different from the control. Statistical significance was indicated by single asterisk (p < 0.05), double asterisks (p < 0.01), and triple asterisks (p < 0.001).
Results
NGF induces global reversal of trimethylated H3K9
Our previous study showed that NGF treatment of PC12h cells resulted in a temporal upregulation of both dor promoter activity and mRNA levels [11]. We hypothesize that NGF induces epigenetic modifications of histone H3 tails to temporally induce dor (and possibly other late phase genes) during differentiation. To test this hypothesis, we carried out a series of experiments. Acetylation of H3K9 is associated with euchromatin gene activation, while methylation of H3K9 is associated with the gene repression locus [25]. Thus, we used Western blot analyses to examine the status of H3K9me3 and acetylated H3K9 (H3K9ac). NGF treatment led to an approximately 40% decrease in the global level of H3K9me3 at 24 h (p < 0.001) (Fig. 1). Dimethylated H3K9 (H3K9me2) had a spurt increase at 30 min and then a slow decrease after 4 h (Fig. 1). In contrast, the levels of H3K9ac slightly increased at 4 h and then leveled off (Fig. 1). To our knowledge, this is the first experimental evidence to show that NGF signaling selectively and globally reverses H3K9 trimethylation. These preliminary results prompted us to investigate further the locus-specific trimethylation of H3K9 in vivo.
Fig. 1.
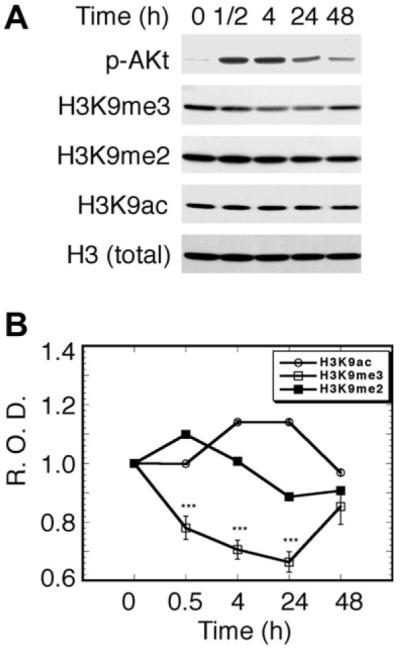
Effect of NGF on global modifications of histone H3. (A) Western blot analysis of NGF-induced methylation, demethylation, and acetylation of H3K9. Cells were treated with NGF at the indicated times in the presence of 0.1% calf serum. p-Akt, phosphorylated Akt (Ser 473), which can represent PI3K kinase activity; H3, total histone H3 (used as a control for normalizing the optical density of each band); H3K9ac, acetylated Lys9 of histone H3; H3K9me2, dimethylated Lys9 of histone H3; H3K9me3, trimethylated Lys9 of histone H3. Each protein band was identified by the corresponding specific antibody (see Materials and methods). (B) Semi-quantitative presentation of H3K9 modifications. The optical density of each band was divided by that of the H3 band to give the relative optical density (R.O.D.), which was further normalized to be one unit at time zero. Data are means ± SEM of three measurements (*** p < 0.001).
Dynamic demethylation of H3K9me3
H3K9 methylation has been linked to the formation and maintenance of repressive heterochromatin [26,27]. To examine the modification of histones in the putative dor promoter region, we first evaluated the trimethylation status of H3K9. To examine the effect of NGF on the trimethylation of H3K9, we evaluated the association of H3K9me3 with the dor promoter region in vivo using ChIP assays. NGF treatment resulted in a dynamic change in the association of H3K9me3 with the dor gene, illustrated by semi-quantitative PCR (Fig. 2A) and quantified by quantitative real-time PCR (qPCR) (Fig. 2B).
Fig. 2.
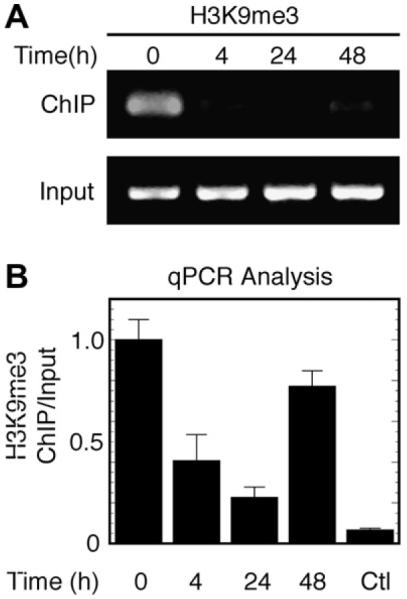
Dynamic regulation of trimethylated H3K9 on the dor gene. (A) Time-dependent NGF-mediated reversal of trimethylated H3K9. (B) Quantitative real-time PCR analysis of H3K9me3 association. After cells were treated with NGF at the indicated times in the conditioned medium (see Materials and methods), cells were harvested and trimethylated H3K9-bound chromatin DNAs were immunoprecipitated. H3K9me3, trimethylated H3K9; ChIP, chromatin immunoprecipitation; input, DNA before immunoprecipitation. ChIP assays were carried out according to the manufacturer’s protocol as described. Ctl, rabbit serum as a negative control. The ratio of ChIP to input DNA at time zero was normalized to be one unit. Data are means ± SEM of three independent experiments in 3–9 real-time PCR replicates.
PI3K signaling-dependent H3K9 trimethylation
To evaluate whether sustained NGF/TrkA/PI3K signaling is required for the epigenetic locus-specific methylation of H3 during the initiation and propagation of transcription, we examined the effects of the PI3K inhibitor LY294002 and the MAPK inhibitor PD98059 on the association of trimethylated H3 with the dor gene. Interestingly, inhibition of PI3K signaling resulted in significant upregulation of H3K9me3 on the dor promoter (*** p < 0.001), while no appreciable effect was observed in the presence of PD98059 (p > 0.05) (Fig. 3). This is consistent with our previous findings that PI3K/Akt signaling is the major pathway required for NGF-induced dor gene expression, and that MAPK signaling plays a less prominent role [11]. The role of NGF/PI3K signaling in reversing the trimethylation of H3K9 indicates that H3K9 trimethylation might be associated functionally with NGF/PI3K survival signaling during NGF-induced differentiation.
Fig. 3.
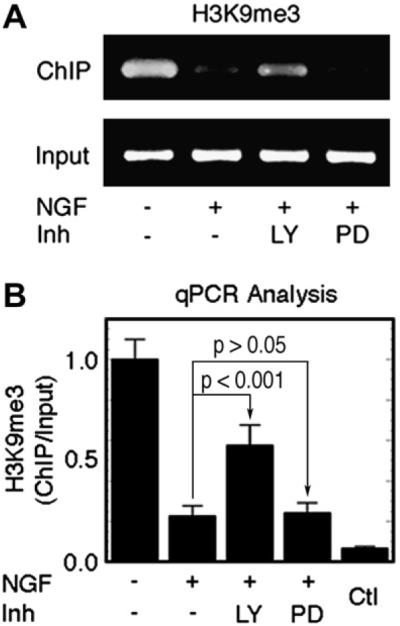
NGF/PI3K-signaling-dependent association of trimethylated H3K9 with the dor gene. (A) Blocking NGF/PI3K signaling resulted in an increase in the H3K9me3 level. The results are representative of three separate semi-quantitative PCR experiments. (B) qPCR analysis of H3K9me3 association with the dor gene. H3K9me3, trimethylated H3K9; LY, LY294002 (20 μM); PD, PD98059 (20 μM); Ctl, rabbit serum as a negative control. PC12h cells were treated with mock control (DMSO), LY, or PD for 24 h. Data are means ± SEM of three independent experiments in 3–11 PCR replicates.
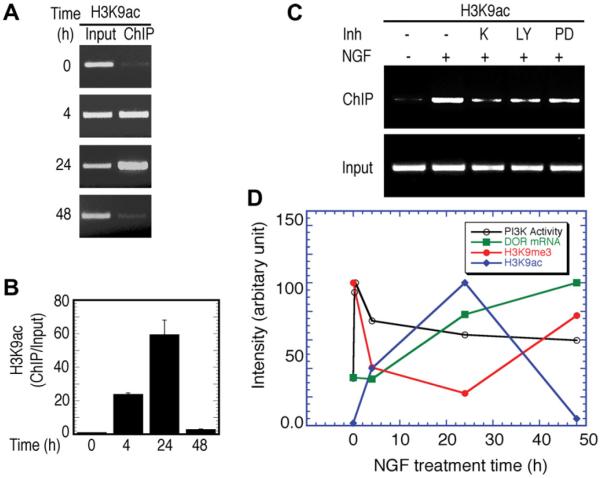
Time-dependent association of acetylated H3K9 with the dor gene
Demethylation of trimethylated H3K9 should provide opportunities for additional acetylation of H3K9 on the same histone tails. NGF treatment of PC12h cells resulted in a time-dependent increase in the association of acetylated H3K9 with dor in vivo (Fig. 4A and B). Furthermore, the association of H3K9ac is dependent upon TrkA and PI3K kinase signaling and, to a much less degree, upon MAPK signaling, as illustrated by studies using their corresponding inhibitors (K252, LY294002, and PD98059, respectively) (Fig. 4C). The decrease in the H3K9ac levels after 24 h of NGF treatment suggests that acetylation of H3K9 may only play a role in the initiation, but not in the termination, of transcription.
Fig. 4.
Correlation of histone H3K9 modifications with the temporal induction of dor mRNA during NGF-induced differentiation. (A) Time-dependent NGF-induced association of H3K9ac with the dor gene. (B) qPCR analysis of ChIP DNA. Data are means ± SEM of triplicate experiments. (C) Effect of kinase inhibitors on H3K9ac association. The representative results from the semi-quantitative ChIP-PCR analysis are shown in A and C. K, K252a (50 nM; an NGF receptor TrkA inhibitor); LY, LY294002 (20 μM); PD, PD98059 (20 μM). (D) Correlation of histone H3K9 modifications with the temporal induction of dor mRNA during NGF-induced differentiation. The dor mRNA data points were extracted from a time-dependent induction of dor mRNA experiment which is similar to the one that previously published [11]. PI3K activity, presented by the level of phosphorylated Akt, which was extracted from the experiment that previously published [11]. H3K9ac, acetylated Lys9 of histone H3 from qPCR analysis (B); H3K9me3, trimethylated Lys9 of histone H3 from qPCR analysis (Fig. 2B). The maximum arbitrary unit at the y-axis for each corresponding variable is normalized to be 100% for comparison purposes.
Discussion
The dynamic trimethylation reversal and acetylation of histone H3 correlate well with the kinetics of NGF/PI3K-signaling-dependent dor expression (Fig. 4D, [11]). These results indicate that PI3K signaling could play a critical role in both the initiation and propagation of dor transcription. This is also consistent with our previous finding that sustained NGF/PI3K signaling is important for the levels of the rat dor mRNA and the activity of the mouse dor promoter [11]. Based on our results, a possible mechanism of NGF-induced late phase dor gene expression has emerged: NGF treatment provides sustained activation of NGF/PI3K signaling, which then results in a series of well-coordinated biochemical and cellular events. Activation of PI3K/Akt signaling resulted in translocalization of NF-κB from the cytosol to the nucleus [11], which is then associated with p300 in the nucleus [28]. Moreover, the NF-κB complex in the nucleus is also associated with the mouse dor promoter in living cells [29]. PI3K signaling then activates a yet to be identified demethylase, which then reversal of H3K9me3, and providing a free H3K9 to be acetylated. Acetylation of H3K9 is most likely accomplished by p300 because p300 can be associated with activated NF-κB complex in the nucleus [28]. Reversal of H3K9 trimethylation likely precedes acetylation of H3K9 on the same histone tails. Eventually, the locus-specific acetylation of H3K9 (together with other modifications by yet-to-be-identified coactivators/chromatin modifying factors) helps to initiate NGF-induced gene transcription. The mechanism proposed is also consistent with the consensus that H3K9 methylation is generally involved in transcriptional repression [25,27], although exceptions to this phenomenon have been observed [30]. The reversal of H3K9 trimethylation in NGF-induced differentiation of PC12 cells in this study, and the inducibility of H3K9 methylation in lipopolysaccharide-treated immune cells [31] together supports the notion that the regulation of H3K9 trimethylation can have broad implications in the expression of NGF-induced genes.
In this study, we observed the well-coordinated temporal methylation and acetylation of H3K9. These modifications underscore an epigenetic regulatory mechanism for controlling survival-associated dor expression during neuronal differentiation. Indeed, dor is expressed both temporally and spatially during neuronal development[12,13]. Pharmacological studies have demonstrated that the delta opioid receptor signaling plays an important role in NGF-induced neuronal differentiation in PC12 cells [5]. Moreover, H3K9 methylation has been directly associated with DNA CpG methylation, e.g., H3K9 methylation is necessary for DNA methylation to occur in Neurospora crassa [26]. The previous studies have shown that in the unstimulated mouse neuroblastoma NS20Y and neuro2A cell lines that the levels of dor gene expression correlate with those of DNA CpG methylation in the mouse dor promoter region [32]. NGF-induced differentiation of PC12 cells has been recently reported to be associated with DNA methyltransferase3B [33]. Our current results demonstrate that NGF induces both global and locus-specific reversal of H3K9me3, suggesting that NGF/PI3K signaling might be involved in mediating H3K9me3-specific demethylase activity. Hence, H3K9 trimethylation could be a key linkage between histone methylation and DNA CpG methylation, providing an epigenetic repressive control mechanism for dor gene expression. Indeed, H3K9me3 demethylation may represent a general mechanism for temporal NGF induction of late phase genes during neuronal development through locus-specific epigenetic chromatin remodeling.
In summary, this study shows for the first time that dynamic modification of H3K9 is NGF/PI3K-signaling-dependent and associated with temporal expression of the dor gene during NGF-induced differentiation of PC12 cells. Our findings provide a new epigenetic regulatory mechanism to explain how NGF induces the late phase dor gene in the PC12 cell model. Our results suggest a role for PI3K signaling in the regulation of the status of H3K9me3 during NGF-induced neuronal differentiation. To our knowledge, this is the first demonstration that NGF/PI3K-survival-signaling selectively regulates the trimethylation status of H3K9. Because both NGF/PI3K signaling and opioid signaling are involved in cell survival, neuronal differentiation, and development, reversal of trimethylated H3K9 may play an important role in the opioid receptor-mediated effectors.
Acknowledgments
This work was supported by National Institute of Health Grants DA-000546, DA-001583, DA-001806, DA07339, KO5-DA-70554 (HHL), K05-DA00513 (PYL), and by A. & F. Stark Fund of the Minnesota Medical Foundation.
References
- [1].Kieffer BL, Gaveriaux-Ruff C. Exploring the opioid system by gene knockout. Prog. Neurobiol. 2002;66:285–306. doi: 10.1016/s0301-0082(02)00008-4. [DOI] [PubMed] [Google Scholar]
- [2].Bigliardi-Qi M, Gaveriaux-Ruff C, Zhou H, Hell C, Bady P, Rufli T, Kieffer B, Bigliardi P. Deletion of delta-opioid receptor in mice alters skin differentiation and delays wound healing. Differentiation. 2006;74:174–185. doi: 10.1111/j.1432-0436.2006.00065.x. [DOI] [PubMed] [Google Scholar]
- [3].Zhang J, Gibney GT, Zhao P, Xia Y. Neuroprotective role of delta-opioid receptors in cortical neurons. Am. J. Physiol. Cell Physiol. 2002;282:C1225–C1234. doi: 10.1152/ajpcell.00226.2001. [DOI] [PubMed] [Google Scholar]
- [4].Narita M, Kuzumaki N, Miyatake M, Sato F, Wachi H, Seyama Y, Suzuki T. Role of delta-opioid receptor function in neurogenesis and neuroprotection. J. Neurochem. 2006;97:1494–1505. doi: 10.1111/j.1471-4159.2006.03849.x. [DOI] [PubMed] [Google Scholar]
- [5].Chang SF, Mok MS. The influence of different sub-type delta opioid receptors in nerve growth factor-induced neuronal differentiation in rat pheochromocytoma PC12 cell. Neurosci. Lett. 2001;314:29–32. doi: 10.1016/s0304-3940(01)02280-7. [DOI] [PubMed] [Google Scholar]
- [6].Dermitzaki E, Chatzaki E, Gravanis A, Margioris AN. Opioids transiently prevent activation of apoptotic mechanisms following short periods of serum withdrawal. J. Neurochem. 2000;74:960–969. doi: 10.1046/j.1471-4159.2000.0740960.x. [DOI] [PubMed] [Google Scholar]
- [7].Sofroniew MV, Howe CL, Mobley WC. Nerve growth factor signaling, neuroprotection, and neural repair. Annu. Rev. Neurosci. 2001;24:1217–1281. doi: 10.1146/annurev.neuro.24.1.1217. [DOI] [PubMed] [Google Scholar]
- [8].Greene LA, Tischler AS. Establishment of a noradrenergic clonal line of rat adrenal pheochromocytoma cells which respond to nerve growth factor. Proc. Natl. Acad. Sci. USA. 1976;73:2424–2428. doi: 10.1073/pnas.73.7.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Abood ME, Tao Q. Characterization of a delta opioid receptor in rat pheochromocytoma cells. J. Pharmacol. Exp. Ther. 1995;274:1566–1573. [PubMed] [Google Scholar]
- [10].Inoue N, Hatanaka H. Nerve growth factor induces specific enkephalin binding sites in a nerve cell line. J. Biol. Chem. 1982;257:9238–9241. [PubMed] [Google Scholar]
- [11].Chen YL, Law PY, Loh HH. Sustained activation of phosphatidylinositol 3-kinase/Akt/nuclear factor {kappa}B signaling mediates G protein-coupled {delta}-opioid receptor gene expression. J. Biol. Chem. 2006;281:3067–3074. doi: 10.1074/jbc.M506721200. [DOI] [PubMed] [Google Scholar]
- [12].Zhu Y, Hsu MS, Pintar JE. Developmental expression of the mu, kappa, and delta opioid receptor mRNAs in mouse. J. Neurosci. 1998;18:2538–2549. doi: 10.1523/JNEUROSCI.18-07-02538.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Georges F, Normand E, Bloch B, Le Moine C. Opioid receptor gene expression in the rat brain during ontogeny, with special reference to the mesostriatal system: an in situ hybridization study. Brain Res. Dev. Brain Res. 1998;109:187–199. doi: 10.1016/s0165-3806(98)00082-0. [DOI] [PubMed] [Google Scholar]
- [14].Huang EJ, Reichardt LF. TRK receptors: roles in neuronal signal transduction. Annu. Rev. Biochem. 2003;27:27. doi: 10.1146/annurev.biochem.72.121801.161629. [DOI] [PubMed] [Google Scholar]
- [15].Clayton AL, Rose S, Barratt MJ, Mahadevan LC. Phosphoacetylation of histone H3 on c-fos- and c-jun-associated nucleosomes upon gene activation. EMBO J. 2000;19:3714–3726. doi: 10.1093/emboj/19.14.3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Cheung P, Tanner KG, Cheung WL, Sassone-Corsi P, Denu JM, Allis CD. Synergistic coupling of histone H3 phosphorylation and acetylation in response to epidermal growth factor stimulation. Mol. Cell. 2000;5:905–915. doi: 10.1016/s1097-2765(00)80256-7. [DOI] [PubMed] [Google Scholar]
- [17].Lo WS, Trievel RC, Rojas JR, Duggan L, Hsu JY, Allis CD, Marmorstein R, Berger SL. Phosphorylation of serine 10 in histone H3 is functionally linked in vitro and in vivo to Gcn5-mediated acetylation at lysine 14. Mol. Cell. 2000;5:917–926. doi: 10.1016/s1097-2765(00)80257-9. [DOI] [PubMed] [Google Scholar]
- [18].Lee NH, Weinstock KG, Kirkness EF, Earle-Hughes JA, Fuldner RA, Marmaros S, Glodek A, Gocayne JD, Adams MD, Kerlavage AR, et al. Comparative expressed-sequence-tag analysis of differential gene expression profiles in PC-12 cells before and after nerve growth factor treatment. Proc. Natl. Acad. Sci. USA. 1995;92:8303–8307. doi: 10.1073/pnas.92.18.8303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Wong K, Zhang J, Awasthi S, Sharma A, Rogers L, Matlock EF, Van Lint C, Karpova T, McNally J, Harrod R. Nerve growth factor receptor signaling induces histone acetyltransferase domain-dependent nuclear translocation of p300/CREB-binding protein-associated factor and hGCN5 acetyltransferases. J. Biol. Chem. 2004;279:55667–55674. doi: 10.1074/jbc.M408174200. [DOI] [PubMed] [Google Scholar]
- [20].Middleton G, Wyatt S, Ninkina N, Davies AM. Reciprocal developmental changes in the roles of Bcl-w and Bcl-x(L) in regulating sensory neuron survival. Development. 2001;128:447–457. doi: 10.1242/dev.128.3.447. [DOI] [PubMed] [Google Scholar]
- [21].Brunet A, Datta SR, Greenberg ME. Transcription-dependent and -independent control of neuronal survival by the PI3K-Akt signaling pathway. Curr. Opin. Neurobiol. 2001;11:297–305. doi: 10.1016/s0959-4388(00)00211-7. [DOI] [PubMed] [Google Scholar]
- [22].Yuan J, Yankner BA. Apoptosis in the nervous system. Nature. 2000;407:802–809. doi: 10.1038/35037739. [DOI] [PubMed] [Google Scholar]
- [23].Chen YL, Law PY, Loh HH. Inhibition of akt/protein kinase B signaling by naltrindole in small cell lung cancer cells. Cancer Res. 2004;64:8723–8730. doi: 10.1158/0008-5472.CAN-03-3091. [DOI] [PubMed] [Google Scholar]
- [24].Augustin LB, Felsheim RF, Min BH, Fuchs SM, Fuchs JA, Loh HH. Genomic structure of the mouse delta opioid receptor gene. Biochem. Biophys. Res. Commun. 1995;207:111–119. doi: 10.1006/bbrc.1995.1160. [DOI] [PubMed] [Google Scholar]
- [25].Lachner M, Jenuwein T. The many faces of histone lysine methylation. Curr. Opin. Cell Biol. 2002;14:286–298. doi: 10.1016/s0955-0674(02)00335-6. [DOI] [PubMed] [Google Scholar]
- [26].Bannister AJ, Kouzarides T. Reversing histone methylation. Nature. 2005;436:1103–1106. doi: 10.1038/nature04048. [DOI] [PubMed] [Google Scholar]
- [27].Martin C, Zhang Y. The diverse functions of histone lysine methylation. Nat. Rev. Mol. Cell Biol. 2005;6:838–849. doi: 10.1038/nrm1761. [DOI] [PubMed] [Google Scholar]
- [28].Zhong H, May MJ, Jimi E, Ghosh S. The phosphorylation status of nuclear NF-kappa B determines its association with CBP/p300 or HDAC-1. Mol. Cell. 2002;9:625–636. doi: 10.1016/s1097-2765(02)00477-x. [DOI] [PubMed] [Google Scholar]
- [29].Chen YL, Law PY, Loh HH. Action of NF-kappaB on the delta opioid receptor gene promoter. Biochem. Biophys. Res. Commun. 2007;352:818–822. doi: 10.1016/j.bbrc.2006.11.103. [DOI] [PubMed] [Google Scholar]
- [30].Vakoc CR, Mandat SA, Olenchock BA, Blobel GA. Histone H3 lysine 9 methylation and HP1gamma are associated with transcription elongation through mammalian chromatin. Mol. Cell. 2005;19:381–391. doi: 10.1016/j.molcel.2005.06.011. [DOI] [PubMed] [Google Scholar]
- [31].Saccani S, Natoli G. Dynamic changes in histone H3 Lys 9 methylation occurring at tightly regulated inducible inflammatory genes. Genes Dev. 2002;16:2219–2224. doi: 10.1101/gad.232502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Wang G, Liu T, Wei LN, Law PY, Loh HH. DNA methylation-related chromatin modification in the regulation of mouse delta-opioid receptor gene. Mol. Pharmacol. 2005;67:2032–2039. doi: 10.1124/mol.105.011056. [DOI] [PubMed] [Google Scholar]
- [33].Bai S, Ghoshal K, Jacob ST. Identification of T-cadherin as a novel target of DNA methyltransferase 3B and its role in the suppression of nerve growth factor-mediated neurite outgrowth in PC12 cells. J. Biol. Chem. 2006;281:13604–13611. doi: 10.1074/jbc.M513278200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]