Abstract
A comprehensive re-evaluation of the G protein alpha subunit genes specifically expressed in taste buds in the tongue epithelium of rodents revealed that Gq and G14 of the Gq class and Gi2 and Ggust (Gt3, also known as gustducin) of the Gi class are expressed in mammalian taste buds. Meanwhile, a database search of fish genomes revealed the absence of a gene encoding an ortholog of the mammalian Ggust gene, which mediates sweet, umami, and bitter taste signals in mammalian taste receptor cells (TRCs). Histochemical screening identified two G protein alpha subunit genes, zfGia and zfG14, expressed in subsets of TRCs in zebrafish. The expression patterns of zfGia and zfG14 in taste buds were mutually exclusive, and the expression of known T1R and T2R genes in zebrafish was restricted to a subset of zfGia-expressing TRCs. These findings highlight the existence of a novel subset of TRCs in zebrafish that is absent in mammals and suggest that unidentified G protein-coupled receptors are expressed in zfG14-expressing TRCs and in zfGia-expressing TRCs where known T1R and T2R genes were not expressed in zebrafish. The existence of not only generalized but also specialized subsets of TRCs may imply a strong connection between the evolution of the peripheral gustatory system and the evolution of particular species.
Keywords: taste bud, taste receptor cell, G protein, signal transduction, zebrafish, mouse
Introduction
Heterotrimeric G proteins transduce extracellular stimuli received by G protein coupled receptors (GPCRs) to intracellular signaling cascades and are involved in a variety of physiological functions. Disruption of the activities of G proteins leads to physiological disorders and diseases (Downes and Gautam, 1999). Some G proteins, especially alpha subunits, exhibit specific spatial expression patterns and are directly involved in the specific functions of individual tissues and/or cells (Wettschureck and Offermanns, 2005). Therefore, identifying and characterizing the types of G proteins in a given tissue and/or cell could provide important insight into the functions of that tissue and/or cell.
Taste is a chemical sense closely related to food intake behavior. Vertebrates detect chemicals in food using specialized epithelial sensory cells, termed taste receptor cells (TRCs), derived from the local epithelium (Barlow and Northcutt, 1995; Stone et al., 1995). Dozens of taste receptor cells unite to form a taste bud, a gustatory apparatus distributed in oral, pharyngeal, and laryngeal (or esophageal in fish) epithelia in vertebrates. In mammals, individual taste buds are composed of functionally different TRCs (Chandrashekar et al., 2006), including T1R-expressing cells responsible for sweet and umami tastes (Zhao et al., 2003) and T2R-expressing cells responsible for bitter taste (Mueller et al., 2005), both of which utilize common molecules, such as phospholipase C-β2 (PLC-β2) and transient receptor potential channel M5 (TRPM5), in their downstream intracellular signaling cascades (Zhang et al., 2003). The expression of G protein alpha subunit (Gna) genes in taste buds has been shown mainly by RT-PCR, a technique that is somewhat biased toward positive results for gene expression. Histochemical analyses, on the other hand, have revealed the expression of Ggust, G15, Gq, Gi2, and G14 in a subset of taste bud cells (Kusakabe et al., 1998; Kusakabe et al., 2000; McLaughlin et al., 1992; Shindo et al., 2008; Tizzano et al., 2008). These results suggest that the Gq and Gi classes are involved in taste signaling in TRCs. Knockout mice for Ggust have demonstrated the involvement of Ggust in sweet, umami, and bitter taste reception, at least in part. Molecular data regarding T1Rs, T2Rs, PLC-β2, and TRPM5 in fish suggest that there are common mechanisms of taste reception and intracellular signaling among vertebrates (Ishimaru et al., 2005; Oike et al., 2007; Yasuoka et al., 2004; Yoshida et al., 2007). Fish genomes, however, show species-specific divergence of the T1R2 genes (Ishimaru et al., 2005). There are some differences between the fish and mammalian gustatory systems, likely due to evolutionary divergence.
To elucidate the molecular mechanisms of taste reception in fish and obtain insight into the phylogenetic differentiation of TRCs in vertebrates, we carried out a search for the Gna genes expressed in TRCs in fish. Our results showed that there is no ortholog of rodent Ggust in fish or amphibian genomes and that two Gna genes, zfGia and zfG14, are expressed in zebrafish taste buds in a mutually exclusive manner. Interestingly, the expression of the zfT1R and zfT2R genes identified thus far was restricted to a subset of zfGia-expressing TRCs, suggesting the existence of novel TRC subsets and GPCRs in zebrafish taste buds.
Materials and Methods
Phylogenetic analysis of G protein alpha subunits
Full-length amino acid sequences of mammalian Gna genes were used to search for similar genes in vertebrate species in the public database (National Center for Biotechnology Information, NCBI) using the TBLASTN program (NCBI). The amino acid sequences of zebrafish G protein alpha subunits were used as queries to search for similar proteins in medaka (MEDAKA1 assembly), fugu (FUGU 4.0 assembly), and puffer fish (TETRAODON 8.0 assembly) in the Ensembl genome database (http://www.ensembl.org/). Full-length amino acid sequences were aligned using the FFT-NS-i program on the MAFFT version 6 Web site (http://align.bmr.kyushu-u.ac.jp/mafft/online/server/index.html) (Katoh et al., 2002) with default settings. The alignments were examined, and some predicted sequences obtained from the Ensembl database were manually edited based on the gene structure of the orthologous genes. All gap sites in the alignment were excluded. Phylogenetic analyses were performed using maximum likelihood (ML) and neighbor-joining (NJ) methods. The ML tree was built with Proml from the Phylip3.69 package (http://evolution.genetics.washington.edu/phylip.html) using the JTT model and the global rearrangement option, and bootstrap values calculated with 100 replicates were assigned to the tree. The NJ tree building and bootstrap analysis with 1000 replicates were performed with MEGA4 software (Tamura et al., 2007) using the JTT model.
DNA microarray analysis
DNA microarray experiment with Rat Genome 230 2.0 (Affymetrix, Santa Clara, CA) was performed according to the manufacturer’s protocol, and gene expression data were obtained as described previously (Ohmoto et al., 2006). The values indicating expression level were normalized relative to that of GAPDH (GenBank accession no. NM_017008) to 7,000.
Cloning of cDNA and DNA fragment of coding region
Total RNA was extracted using TRIzol (Invitrogen, San Diego, CA) from the brain and circumvallate papillae of C57BL6/J mice or from the brain and gills of adult zebrafish (Danio rerio) purchased from a commercial supplier. First-strand cDNA was synthesized using oligo-dT primers and the SuperScript II reverse transcriptase (Invitrogen) and used as a template for RT-PCR. To obtain the intronless coding region of zebrafish T2Rs or V1Rs genes, genomic DNA was used as a template. The amplified cDNA fragments were cloned into Bluescript II SK- (Stratagene, La Jolla, CA) and sequenced using a 3130 automated DNA sequencer (Applied Biosystems Inc., Foster City, CA). The probe regions in the zebrafish Gna and other genes are shown in Table 1 and Table 2, respectively. Plasmids containing cDNA fragment of zebrafish ORs, V2Rs, and TAAR9 were kindly gifted by Dr. Y. Yoshihara and Dr. Y. Sato of RIKEN Brain Science Institute (Sato et al., 2005; Sato et al., 2007).
Table 1.
Information on G protein alpha subunit gene and probe regions.
| Gene Name | Gene Symbol | Accession No. | Probe Region |
|---|---|---|---|
| zfG11-1 | zgc:101761 | BC085433 | 121-2209 |
| zfG11-2 | gna11 | NM_001045036 | 1-1310 |
| zfGq | gnaq | XM_693545 | 1-1080* |
| NW_001513461 | 21253-22300* | ||
| zfG14 | gna14a | XM_678897 | 1-1065 |
| zfG14L | gna14 | NM_001003753 | 27-1682 |
| zfGi1 | gnai1 | NM_200971 | 267-1791 |
| zfGia | gnaia | NM_198805 | 375-1863 |
| zfGi2 | gnai2 | NM_199842 | 1-1142 |
| zfGi2L | gnai2l | NM_110111818 | 403-1657 |
| zfGi3 | gnai3 | XM_001343090 | 103-1527 |
Probe region of zfGq used for in situ hybridization analysis corresponds to the total of 1080 bp of XM_693545 (nucleotide number 1-1080) and 1053 bp of NW_001513461 (21253-22300).
Table 2.
Information on probe region of mouse and zebrafish genes.
| Gene Name | Accession No. | Probe Region | Chr. |
|---|---|---|---|
| mGq | NM_008139 | 481-2245 | |
| mG14 | NM_008137 | 651-3193 | |
| mGi2 | NM_008138 | 179-2120 | |
| mGgust | AK040065 | 41-1019 | |
| mT1R1 | AF337040 | 765-2761 | |
| mT1R2 | AF337041 | 641-2804 | |
| mT1R3 | AF337039 | 525-2725 | |
| mTRPM5 | AF228681 | 310-3491 | |
| zfPLC-β2 | XR_030066 | 1-2643#1 | |
| zfT1R2a | NM_001039831 | 48,741,431-48,751,664#2, #3 | 8 |
| zfT1R2b | NM_001083856 | 48,813,526-48,823,054#2, #3 | 8 |
| ztT1R3 | NM_001039628 | 42,207,615-42,218,652#2, #3 | 11 |
| zfT2R1a | NM_001039830 | 31,632,022-31,633,050#2 | 8 |
| zfT2R2a | NM_001083864 | 39,766,083-39,767,088#2 | 9 |
| zfT2R3 | NM_001105135 | 39,780,761-39,781,869#2 | 9 |
| zfT2R4 | NM_001020506 | 24,666,166-24,667,191#2 | 9 |
| zfT2R5 | NM_001100629 | 7,198,115-7,199,179#2 | 8 |
| zfV1R1 | NM_001020504 | 221,032-222,033#2 | 22 |
| zfV1R2 | NM_001098395 | 8,153-9,100#2, #4 | 22 |
| zfTaar9 | BC093335 | 1-1022 |
The length of cDNA fragment we cloned was approximately 2.8 kbp, which contains the sequence not appeard in XR_030066.
The 5′ and/or 3′ flanking sequences to coding region were included. Shown were the location on chromosome from Ensemble genome database Zv9 corresponding to the sequence used as a probe. The number of chromosome on which the gene maps were shown in the adjacent column.
The lengths of probe used were 2,478, 2,472, and 2,553, for zfT1R2a, zfT1R2b, and zfT1R3, respectively.
The location on chromosome for zfV1R2 was from Ensemble Zv8.
Tissue preparation
C57BL6/J mice were sacrificed by cervical dislocation, and oral epithelia containing taste buds were dissected, embedded in O.C.T. Compound (Sakura Finetech, Tokyo, Japan), and frozen in liquid nitrogen. Tissue sections (10 μm thickness) were prepared using a cryostat (CM1900, Leica Microsystems, Wetzlar, Germany). Adult zebrafish were purchased from a commercial supplier. The anterior one-fourth of the body was dissected, fixed overnight with 4% paraformaldehyde (PFA) in PBS (pH 7.4) at 4 °C, dehydrated in methanol for 24 h at −20 °C, decalcified in 0.5 M EDTA in PBS for 24 h at 4 °C, and cryoprotected in 30% sucrose in PBS. These samples were embedded in O.C.T. Compound, frozen in liquid nitrogen, and sectioned transversely at 10μm using a cryostat. The sections from mice and zebrafish were then attached to MAS-coated glass slides (Matsunami Glass, Osaka, Japan). All animal experiments were approved by the Animal Care and Use Committees of the University of Tokyo.
In situ hybridization
The in situ hybridization procedure was described previously (Matsumoto et al., 2001; Ohmoto et al., 2006; Ohmoto et al., 2008). In brief, digoxigenin- and fluorescein-labeled antisense RNAs prepared using RNA labeling mix (Roche Diagnostics, Indianapolis, IN) and an RNA polymerase (Stratagene) were used for hybridization after fragmentation under alkaline conditions to a length of about 150 bases. Fresh-frozen sections (prepared from mice) were fixed with 4% PFA and treated with diethylpyrocarbonate. PFA-fixed sections (prepared from zebrafish) were treated with proteinase K (3 μg/ml) for 10 min, post-fixed with 4% PFA in PBS for 10 min, and then acetylated in 0.1 M triethanolamine containing 0.25% acetic anhydride for 10 min. Sections were prehybridized with salmon sperm DNA for 2 hr at 58 °C, and hybridized with 20–200 ng/ml antisense riboprobe(s) for 40 hr at 58 °C. After hybridization, the sections were washed in 0.2 × SSC at 58 °C and blocked in blocking solution containing 0.5% blocking reagent (Roche Diagnostics). For single labeling, signals were developed using alkaline phosphatase–conjugated anti-digoxigenin antibody (1:500, Roche Diagnostics) and 4-nitro blue tetrazolium chloride/5-bromo-4-chloro-3-indolyl-phosphate as chromogenic substrates. For fluorescent double labeling, biotin–conjugated anti-fluorescein antibody (1:500, Vector Laboratories, Burlingame, CA) followed by ABC solution (Vectastain ABC elite kit, Vector Laboratories) and alkaline phosphatase–conjugated anti-digoxigenin antibody (1:500, Roche Diagnostics) were used in combination with biotin-tyramide (1:50, PerkinElmer, Norwalk, CT) followed by Alexa488-conjugated streptavidin (4 μg/ml, Invitrogen) and HNPP Fluorescent Detection Set (Roche Diagnostics), respectively. Stained images were obtained with an Olympus BX51 microscope (Olympus, Tokyo, Japan) equipped with a DP70 cooled CCD digital camera (Olympus). Fluorescent images were obtained with an FV500 confocal laser scanning microscope (Olympus), and image data were transformed into pseudocolor images using FLUOVIEW (Olympus). Brightness and contrast were adjusted using Adobe Photoshop CS (Adobe Systems, Mountain View, CA, U.S.A.). The sense probes for the genes expressed in zebrafish TRCs yielded no signal (supplementary figure 1).
Results
Expression of genes encoding G protein alpha subunits in taste buds across vertebrates
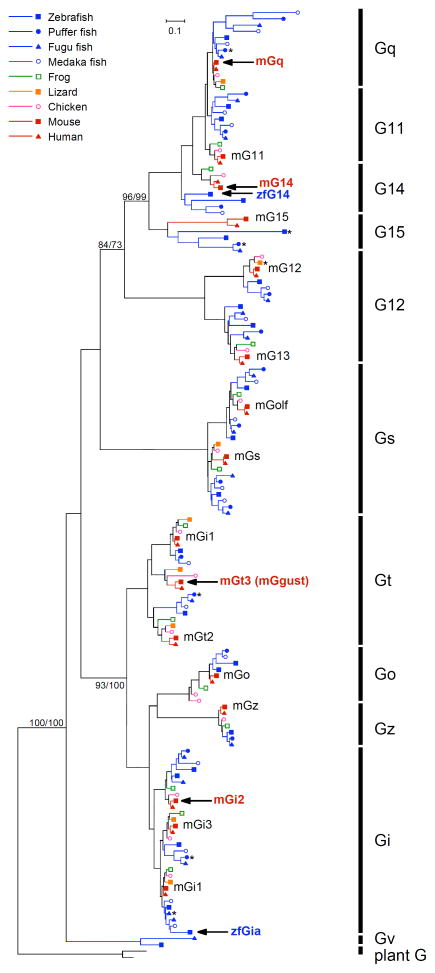
Molecular features of TRCs expressing PLC-β2 are basically conserved well between mammals and fish, while there were some differences about the number of T1R2 and T2R genes. It would be reasonable to speculate that G proteins are expressed in the PLC-β2-expressing TRCs in fish as well as in mammals. Thus, we searched for Gna genes in the database of vertebrate genomes. Interestingly, we failed to find any genes encoding mammalian Ggust orthologs in the frog, zebrafish, fugu, puffer fish, or medaka genomes; however, the fish genomes contained more members of the Gq and Gi classes (e.g., G11, G14, and Gi2) than the mammalian genomes (Fig. 1).
Fig. 1. Unrooted phylogenetic trees of G protein alpha subunits in vertebrates constructed by the maximum likelihood (ML) method.
Plant G alpha subunits are used as an outgroup. Bootstrap values over 70% calculated by the ML and neighbor-joining (NJ) method for branch support were presented in the order of ML/NJ for the major branch. Mouse and zebrafish G alpha genes expressed in taste receptor cells according to the present study and previous studies are shown in bold red and blue letters, respectively, with black arrows. Asterisks indicate genes that have been pseudogenized by insertion or deletion of a single nucleotide that was followed by a downstream in-frame stop codon.
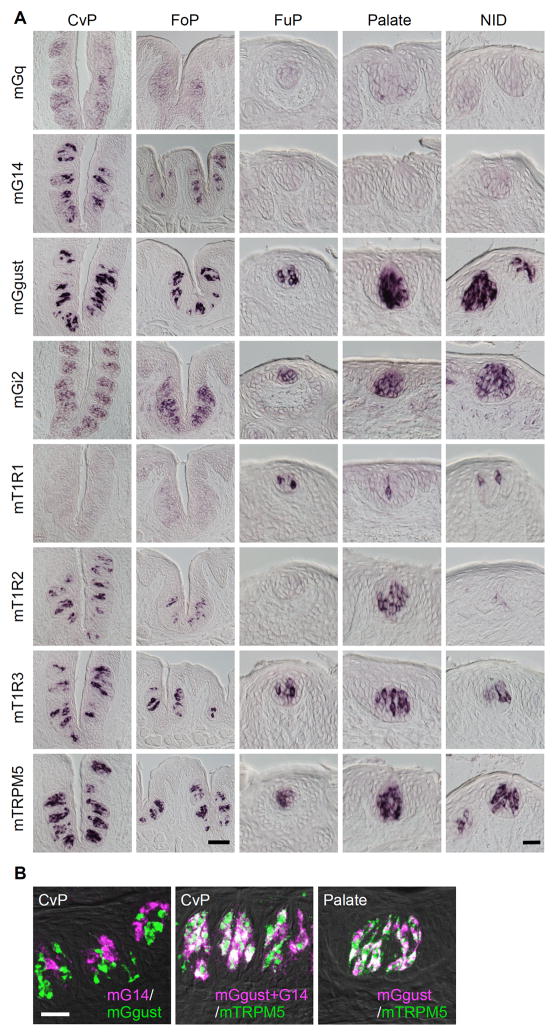
To predict which Gna genes are expressed in fish TRCs, we first conducted comprehensive analysis to know which Gna genes were expressed in TRCs in rodents. Based on gene expression data from isolated taste buds and surrounding tongue epithelia obtained using DNA microarray experiments, which revealed voltage-dependent potassium channel genes specifically expressed in the taste buds of rat circumvallate papillae (Ohmoto et al., 2006), and we found that among all Gna genes identified thus far, Gq, G14, Ggust, and Gi2 are likely to be specifically expressed in taste buds (Table 3). Specific expression of these genes in a subset of taste bud cells was comprehensively re-evaluated by in situ hybridization (Fig. 2A) (Kusakabe et al., 1998; Kusakabe et al., 2000; McLaughlin et al., 1992; Shindo et al., 2008; Tizzano et al., 2008). G15, Gs, and Gi3 are expressed not only in taste buds but also in the surrounding tongue epithelia (Table 3), consistent with our previous histochemical analysis (Kusakabe et al., 2000), although in conflict with our previous immunostaining data for G15 (Kusakabe et al., 1998). The oligopeptide used as an antigen to generate anti-G15 antibody showed a highly specific sequence to rodent G15 species, and it is difficult to consider cross-reactivity to other G protein alpha subunit species like G14 and Ggust, although we do not know why previous immunohistochemistry showed such specific immunoreactivity only to a subset of TRCs. Thus, Gq and G14 of the Gq class and Ggust and Gi2 of the Gi superfamily are expressed in a taste bud-specific manner.
Table 3.
Relative expression level of G protein alpha subunit gene in three regions of tongue epithelium
| TB | TB | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Gene name | Probe Set ID | TB | call#2 | Cvp-Epi | call | Np-Epi | call | Cvp-Epi | Np-Epi | |
| Gq | ||||||||||
| Gnaq#1, *1 | 1375745_at | 1244.3 | P | 487.4 | P | 460.9 | P | 2.55 | 2.70 | |
| Gnaq | 1383055_at | 56.4 | P | 43.8 | P | 35 | P | 1.29 | 1.61 | |
| Gnaq | 1395547_at | 59.7 | P | 40.7 | P | 31.8 | P | 1.47 | 1.88 | |
| Gnaq | 1370091_at | 9.6 | A | 0.8 | A | 1.9 | A | 12.00 | 5.05 | |
| Gna11*2 | 1387822_at | 29.6 | P | 22.2 | P | 33.1 | P | 1.33 | 0.89 | |
| Gna14*3 | 1381557_at | 3415.3 | P | 85.9 | P | 0.4 | A | 39.76 | >100 | |
| Gna14 | 1393825_at | 154.1 | P | 0.9 | A | 0.2 | A | >100 | >100 | |
| Gna15*4 | 1370423_at | 258.5 | P | 444.9 | P | 559.9 | P | 0.58 | 0.46 | |
| G12 | ||||||||||
| Gna12*5 | 1379926_at | 23.4 | A | 3.1 | A | 3.6 | A | 7.55 | 6.50 | |
| Gna12 | 1369278_at | 17.9 | A | 9.7 | A | 19.3 | A | 1.85 | 0.93 | |
| Gna12 | 1389895_at | 1.4 | A | 0.6 | A | 4.5 | A | 2.33 | 0.31 | |
| Gna13 | 1376006_at | 458.7 | P | 626.9 | P | 695.4 | P | 0.73 | 0.66 | |
| Gs | ||||||||||
| Gnas*6 | 1369897_s_at | 2522.2 | P | 1507.1 | P | 1541 | P | 1.67 | 1.64 | |
| Gnas | 1387906_a_at | 55.2 | P | 21.5 | A | 18.8 | A | 2.57 | 2.94 | |
| Gnal (Golf) | 1384229_at | 53.7 | M | 13.1 | A | 20.6 | P | 4.10 | 2.61 | |
| Gnal (Golf) | 1394575_at | 29.4 | A | 15.4 | A | 15.9 | A | 1.91 | 1.85 | |
| Gnal (Golf) | 1382129_at | 6.5 | A | 29.5 | P | 27.6 | P | 0.22 | 0.24 | |
| Gi | ||||||||||
| Gnat1 (Gt-rod) | 1373587_at | 46.2 | A | 58.7 | P | 67.6 | P | 0.79 | 0.68 | |
| Gnat2 (Gt-cone)*7 | 1393840_at | 15.9 | A | 1.4 | A | 1.1 | A | 11.36 | 14.45 | |
| Gnat3 (gustducin)*8 | 1388223_at | 4074.1 | P | 241.3 | P | 7.2 | A | 16.88 | >100 | |
| Gnao | 1375902_at | 17 | A | 14.9 | A | 3.5 | A | 1.14 | 4.86 | |
| Gnao | 1368879_a_at | 6.2 | A | 11.9 | A | 11.8 | P | 0.52 | 0.53 | |
| Gnao | 1377514_at | 4.6 | A | 4 | A | 27 | P | 1.15 | 0.17 | |
| Gnao | 1374314_at | 1.2 | A | 0.3 | A | 0.5 | A | 4.00 | 2.40 | |
| Gnaz | 1368185_at | 4.8 | A | 2.8 | A | 3.2 | A | 1.71 | 1.50 | |
| Gnaz | 1387095_at | 18 | A | 1 | A | 9.3 | P | 18.00 | 1.94 | |
| Gnai1*9 | 1387505_at | 1.1 | A | 4.9 | A | 17.4 | P | 0.22 | 0.06 | |
| Gnai2*10 | 1367844_at | 261.2 | P | 70.6 | P | 75.2 | P | 3.70 | 3.47 | |
| Gnai3*11 | 1368029_at | 1815.7 | P | 3361.2 | P | 4468.7 | P | 0.54 | 0.41 | |
| Gnai3 | 1368030_at | 55 | P | 62.2 | P | 143.8 | P | 0.88 | 0.38 |
The data in bold type indicates genes spedifically expressed in taste buds in the oral epithelium as revealed by in situ hybridization analysis.
Affymetrix software GCOS calculated the possibility of the expression and called P (present), M (marginally present), and A (absent).
*1–*11 Expression in the tongue epithelium containing taste buds was shown by RT-PCR, subsequent cloning, and/or immunohistochemical analyses.
Fig. 2. Comprehensive re-evaluation of the taste bud-specific expression of Gq, G14, Ggust, and Gi2 mRNA in mouse oral epithelia.
The expression of mouse mRNA was revealed by in situ hybridization. A, The expression of mGq and mG14 was restricted to the taste buds in the posterior gustatory papillae, whereas mGi2 and mGgust were expressed in most taste buds in the oral cavity. mT1R1 and mT1R2 were used as references for region-specific expression. mT1R3 and mTRPM5 were used as references for subset-specific expression in taste buds. CvP, circumvallate papilla; FoP, foliate papilla; FuP, fungiform papilla; NID, nasoincisor duct. Scale bars: CvP and FoP (in the bottom panel of FoP), 50 μm; FuP, Palate, and NID (in the bottom panel of NID), 20 μm. B, Double-labeled fluorescent in situ hybridization was carried out to examine the relationship between the expression patterns of mGgust, mG14, and mTRPM5. Summation of signals of mGgust and mG14 were completely overlapped with the signals of mTRPM5 in the CvP where mG14 was expressed. In the palate where mG14 was not expressed, the signals of mGgust and those of mTRPM5 were completely overlapped. n=2 or 3, number of sections ≥4 for CvP, FoP, and NID, and ≥20 for FuP and Palate. Scale bar: 10 μm.
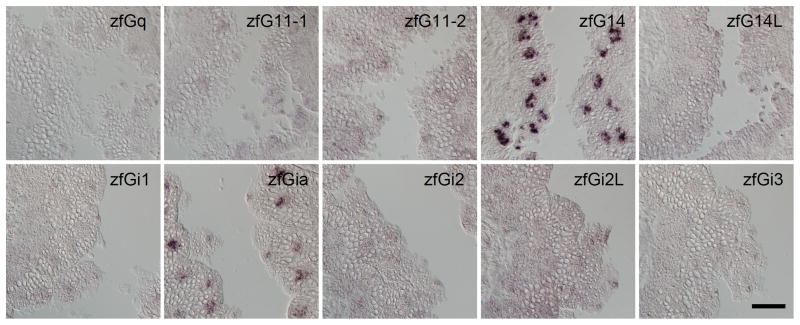
Based on the data from our comprehensive analysis of Gna genes expressed in rodent taste buds, we hypothesized that the Gna gene(s) expressed in fish taste buds belong to the Gq or Gi class. Considering the redundant expression of Gi2 and Ggust in mammalian TRCs and the lack of the Ggust gene in the genomes of teleost fish, it seemed likely that among the Gi superfamily, a gene belonging to the Gi family would be expressed in TRCs in fish. We then carried out in situ hybridization for 10 genes belonging to the Gq class or to the Gi family and found that only G14 (zfG14) of the Gq class and Gia (zfGia) of the Gi family genes were expressed in a subset of taste bud cells in zebrafish (Fig. 3). Compared with other Gna such as Gq and Gi, the fish G14 members ramified from a very early phase of the evolution of Gq class genes (Fig. 1). zfGia is assigned as a member of a small subclade under the node of the Gi1 clade, and zfGia and mfGia contain an extension sequence of approximately 20 amino acid residues at their N termini (Fig. 1). These findings suggest that Gia, which is an ortholog of mammalian Gi1, and G14, both of which are expressed in taste buds, might have evolved in teleost fish genomes in close association with the evolution of teleost fish species.
Fig. 3. Expression of G protein alpha subunits in zebrafish taste buds.
In situ hybridization screening was carried out to identify Gq and Gi species expressed in taste buds in zebrafish. n≥3 (number of sections ≥15) for zfG14 and zfGia, n=2 (no. of sections ≥10) for zfGq, zfG11-1, zfG11-2, and zfG14L, and n=1 (no. of sections ≥5) for zfGi1, zfGi2, zfGi2L, and zfGi3. Scale bar: 50 μm.
Spatial distribution of zfG14 and zfGia in gustatory epithelia
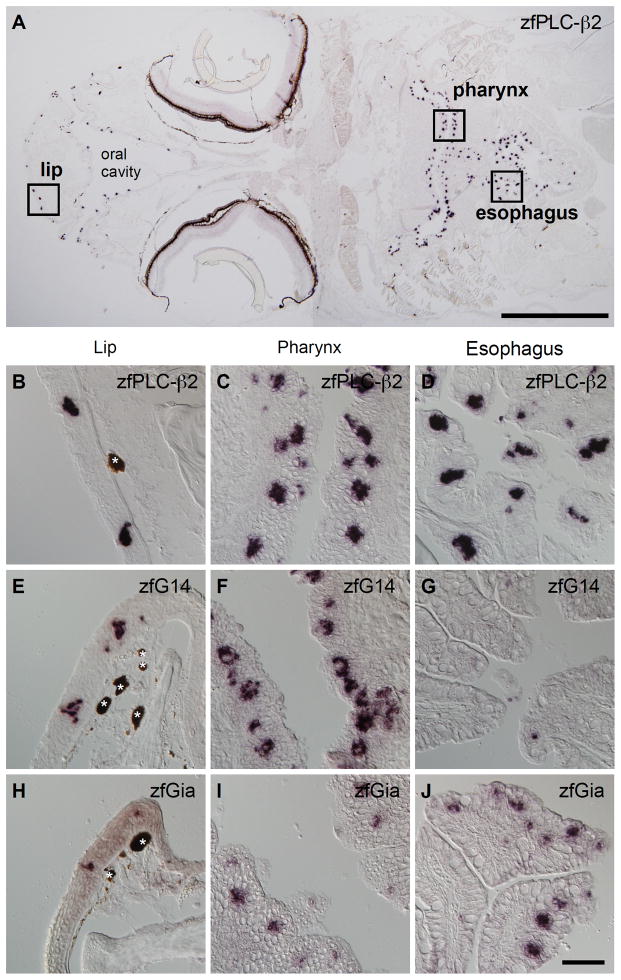
In zebrafish, taste buds are distributed in the lip, oral cavity, pharyngeal region including gill raker (pharynx), and esophagus. We observed strong zfPLC-β2 mRNA expression in taste buds in all of these regions (Fig. 4A–D). The spatial expression patterns of zfGia and zfG14, however, were not so uniform. zfG14 mRNA expression was observed in the lip and pharynx, but rarely in the esophagus (Fig. 4E–G); on the other hand, zfGia expression was observed in the esophagus and pharynx, but rarely in the lip (Fig. 4H–J). The spatially uneven and complementary distributions of zfG14 and zfGia have also been observed for T1R1 and T1R2 in rodents (Fig. 2A) (Hoon et al., 1999), although there is a conflicting report on the segregation of T1R1 and T1R2 expression (Kim et al., 2003).
Fig. 4. Spatial distribution of G14 and Gia mRNAs in zebrafish.
A–D, Distribution of taste buds marked by zfPLC-β2 expression: a global view of a horizontal head section (A) and magnified images of the lip (B), pharyngeal (C), and esophageal (D) epithelia. E–G, Expression of zfG14 mRNA in the lip (E), pharyngeal (F), and esophageal (G) epithelia. H-J, Expression of zfGia mRNA in the lip (H), pharyngeal (I), and esophageal (J) epithelia. mRNA expression revealed by in situ hybridization is indicated by purple staining. Dark brown matter unrelated to this experiment is indicated by white asterisks. Scale bars: A, 1 mm; B–J (in J), 50 μm.
Mutually exclusive expression of zfG14 and zfGia in zfPLC-β2-expressing taste receptor cells
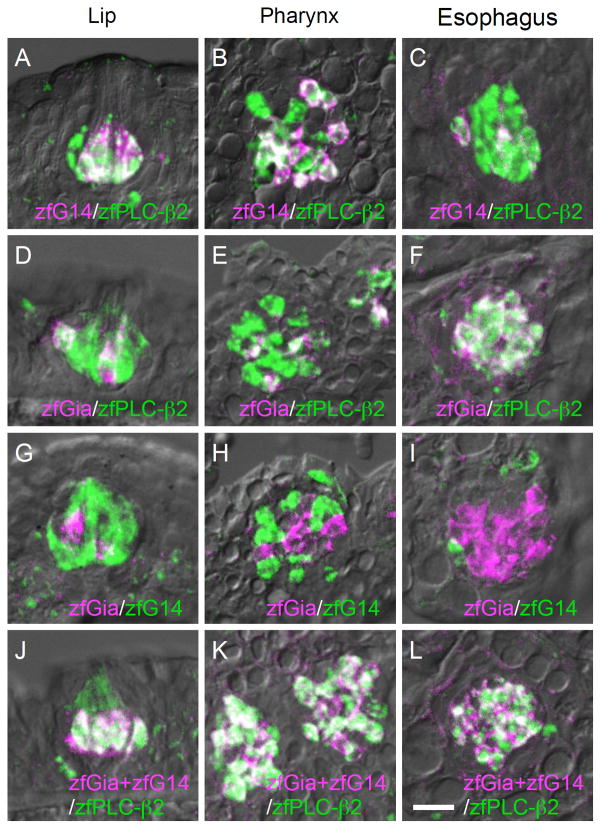
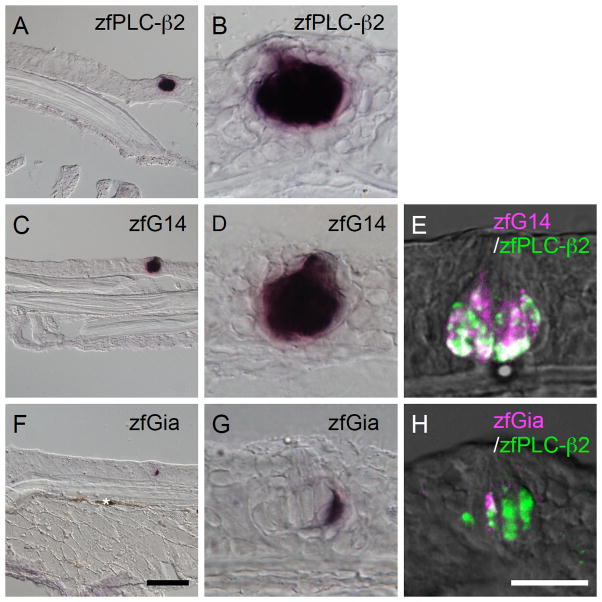
zfG14 and zfGia are likely expressed in PLC-β2-expressing TRCs where known T1R and T2R genes are expressed in zebrafish taste buds. Thus, we carried out double-label fluorescent in situ hybridization analysis and found that both zfG14 and zfGia are expressed in a subset of zfPLC-β2-expressing TRCs (Fig. 5A–F). It is important to note, however, that the zfG14 and zfGia mRNAs were expressed in distinct subsets of TRCs (Fig. 5G–I) and that the summation of the signals from the zfG14 and zfGia mRNAs completely overlapped with the signal from the zfPLC-β2 mRNA (Fig. 5J–L). This is quite similar to the case in rodents, where Ggust and G14 are found in PLC-β2/TRPM5-expressing TRCs in a mutually exclusive manner (Fig. 2B). These findings indicate that zfPLC-β2-expressing TRCs can be classified into two subsets with regard to the expression of G alpha subunits, suggesting that zfG14 and zfGia are involved in intracellular signaling upstream of zfPLC-β2 and zfTRPM5. These results also suggest evolutionary conservation of the physiological function of TRCs expressing G14/PLC-β2/TRPM5 and functional homology between Gia in fish and Ggust in higher vertebrates.
Fig. 5. Relationship of intragemmal expression of G14, Gia, and PLC-β2 in zebrafish gustatory epithelia.
Double-labeled fluorescent in situ hybridization was carried out to examine the relationship between the expression patterns of the following pairs of genes: zfG14 (magenta) and zfPLC-β2 (green) (A–C), zfGia (magenta) and zfPLC-β2 (green) (D–F), zfGia (magenta) and zfG14 (green) (G–I), and zfG14/zfGia (magenta) and zfPLC-β2 (green) (J–L). Signals for zfG14 and zfGia were included in those for zfPLC-β2 (A–F). Signals for zfG14 and zfGia did not overlap (G–I), and their summation was identical to the signal for zfPLC-β2 (J–L), quite similar to the case in mice (Fig. 2B). n=2, number of sections ≥10 for each examination. Scale bars: A–L (in L), 20 μm; M–O (in M), 10 μm.
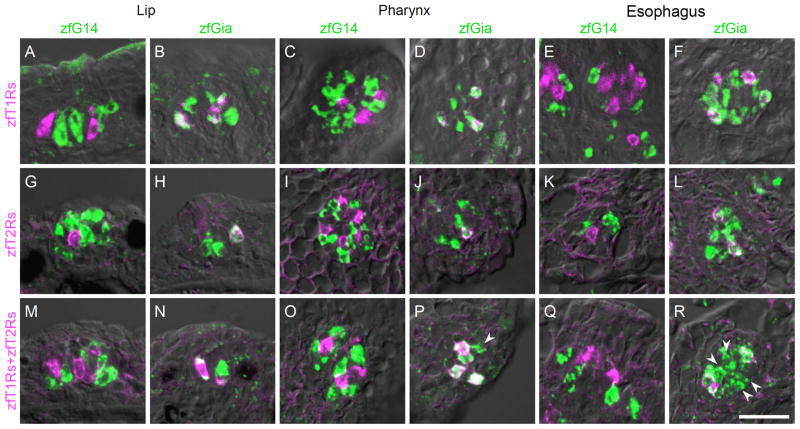
Biased expression of zfT1R and zfT2R in zfGia-expressing taste receptor cells
To understand the relationship between G alpha and taste receptors, we examined the types of receptor genes expressed in zfG14- and zfGia-expressing TRCs. Irrespective of the spatial region, zfT1R genes (zfT1R2a, zfT1R2b and zfT1R3) were expressed in a subset of zfGia-expressing TRCs (Fig. 6A–F), as were zfT2R genes (zfT2R1a, zfT2R2a, zfT2R3, zfT2R4, and zfT2R5) (Fig. 6G–L). These results suggest that zfGia is involved in the signaling pathway downstream of attractive (amino acids) and aversive (denatonium) taste reception by zfT1Rs and zfT2Rs, respectively (Oike et al., 2007). Interestingly, we observed many zfGia-expressing TRCs that lacked both zfT1R and zfT2R expression, especially in the esophagus (Fig. 6P and R, indicated by white arrowheads). These cells represent another subset of zfGia-expressing TRCs. In addition, the zfG14-expressing TRCs did not express any of the taste receptor genes identified to date (Fig. 6M, O, and Q). Unidentified taste receptors other than zfT1Rs and zfT2Rs are likely expressed in the zfG14-expressing TRCs.
Fig. 6. Molecular characterization of taste receptor cells expressing G14 and Gia in zebrafish gustatory epithelia.
Double-labeled fluorescent in situ hybridization was carried out to examine the relationship between the expression of G alpha subunits and the expression of known taste receptors. The probe for zfT1Rs was a mixture of zfT1R2a, zfT1R2b and zfT1R3 riboprobes, which covers all known T1R-expressing TRCs in zebrafish (Ishimaru et al., 2005). The probe for zfT2Rs was also a mixture of zfT2R1a, zfT2R2a, zfT2R3, zfT2R4, and zfT2R5 riboprobes, which covers all known T2R-expressing TRCs in zebrafish (Ishimaru et al., 2005; Oike et al., 2007; Okada et al., 2010). The probe for zfT1Rs+zfT2Rs was a further mixture of the probes for zfT1Rs and zfT2Rs. G14, Gia, and zfT1Rs (A–F); G14, Gia, and zfT2Rs (G–L); and G14, Gia, and all known zebrafish taste receptors (M–R). Both zfT1Rs and zfT2Rs were co-expressed with zfGia (A–L). White arrowheads indicate zfGia-expressing cells that lack zfT1R and zfT2R expression (P and R). n≥2, number of sections ≥10 for each examination. Scale bar: A–R (in R), 20 μm.
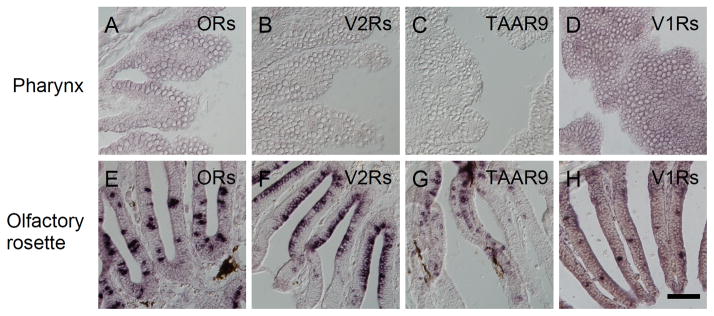
Absence of olfactory GPCRs in taste receptor cells
To identify GPCR(s) expressed in PLC-β2-expressing TRCs where known zfT1Rs and zfT2Rs are not expressed, we examined the expression of olfactory GPCRs in taste buds in zebrafish by in situ hybridization. Mixed probes of OR102-1, OR103-1, OR103-2, OR103-5, OR107-1, and OR119-2; OR111-1, OR111-2, OR111-3, OR111-5, OR111-7, and OR111-10; OR106-11, OR109-9, OR117-1, and OR128-1; and OR101-1, OR124-1, OR125-8, OR133-1, and OR137-1 yielded strong signals in olfactory sensory neurons as reported previously (Sato et al., 2007), but did not yield any signal in taste buds (Fig 7A and E, data not shown). Also, OlfCc1, OlfCd2, OlfCd3 (previously known as VR5.3, VR3.13b, VR3.13a, respectively) (Sato et al., 2005), zfTaar9, zfV1R1, and zfV1R2 mRNAs were not expressed in taste buds, while their abundant expression was observed in the olfactory sensory neurons (Fig. 7B–D and F–H) (Pfister et al., 2007; Pfister and Rodriguez, 2005). In addition, ORA families other than V1R1 and V1R2 families were not expressed in the barbel, lip, or gill, where taste buds are distributed (Saraiva and Korsching, 2007). Thus, taste receptors that would be expressed in PLC-β2-expressing TRCs without expression of known zfT1Rs and zfT2Rs are GPCR(s) other than known olfactory and taste receptors.
Fig. 7. Absence of expression of olfactory GPCRs in zebrafish taste buds.
In situ hybridization screening was carried out to examine the expression of GPCRs in taste buds (A–D) that are known as chemoreceptors expressed in olfactory sensory neurons in zebrafish and/or mammals. Mixed probes for OR111-1, OR111-2, OR111-3, OR111-5, OR111-7, and OR111-10 (A and E); mixed probes for OlfCc1, OlfCd2, and OlfCd3 (previously known as VR5.3, VR3.13b, VR3.13a, respectively) (Sato et al., 2005) (B and F); probe for zfTaar9 (C and G); and mixed probes for zfV1R1 and zfV1R2 (D and H) were used. No GPCR species were expressed in taste buds (A–D), but all probes used yielded strong signals in olfactory epithelium in zebrafish (E–H). n=1, number of sections ≥5 for each examination. Scale bar: 50 μm.
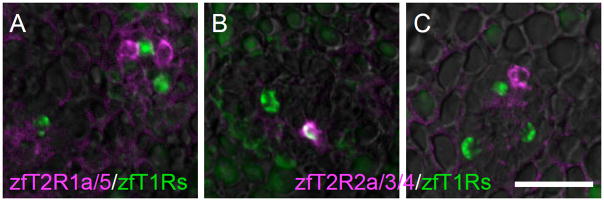
Additional diversity in zfGia-expressing taste receptor cells
As reported previously, zfT2R1a/b and zfT2R5 are expressed in a subset of TRCs distinct from zfT1Rs-expressing TRCs (Fig. 8A). However, the correlation of expression of zfT2R2, 3, and 4 with the expression of zfT1Rs remains to be examined. Using mixed probes for zfT2R2, 3, and 4, we carried out double-label fluorescent in situ hybridization analysis and found that contrary to the case in the mammalian gustatory system, a small population of zfT1Rs-expressing TRCs expressed zfT2R(s) (Fig. 8B and C). This result indicates that TRCs in fish are more diversified than in mammals.
Fig. 8. Relationship of expression of T1Rs and T2Rs in zebrafish taste buds.
Double-labeled fluorescent in situ hybridization was carried out to examine the relationship between the expression patterns of zfT2Rs (magenta) and zfT1Rs (green). The probe for zfT1Rs was a mixture of zfT1R2a, zfT1R2b and zfT1R3 riboprobes, which covers all known T1R-expressing TRCs in zebrafish as described in the legend of Fig. 7. The pattern obtained with the probe mixture for zfT2R1a and zfT2R5 did not overlap with the expression pattern of zfT1Rs (A), consistent with our previous data (Ishimaru et al., 2005; Oike et al., 2007). The probe mixture for zfT2R2a, zfT2R3, and zfT2R4 showed both overlapping expression (B) and distinct expression (C) compared with zfT1Rs expression. n=2, number of sections ≥10 for each examination. Scale bars: 20 μm.
Coexpression of T1R and T2R is observed in solitary chemosensory cells (SCCs) in mammals, in which Ggust, PLC-β2, and TRPM5 are also coexpressed (Finger et al., 2003; Ohmoto et al., 2008). SCCs exist on the body surface in fish (Finger, 1997; Kotrschal et al., 1997), and have been suggested to express amino acid receptor(s), especially an arginine receptor (Finger et al., 1996). Thus, we first examined expression of zfGia, zfG14, and zfPLC-β2 in SCCs. We observed mRNA expression of zfPLC-β2 in the epithelium of the body surface of zebrafish, and its expression was observed in a clustered structure, i.e., taste buds (Fig. 9A and B). zfGia and zfG14 mRNAs were also expressed in the epithelium of the body surface, and their expression was restricted to a subset of zfPLC-β2-expressing clustered cells (Fig. 9C–H). Therefore, we have concluded that zfGia, zfG14, and zfPLC-β2 are expressed in TRCs but not in SCCs. We also examined the expression of V2Rs and T1Rs using mixed zfT1Rs probe and mixed zfV2Rs probe containing zf5.24 and OlfCs, but were unable to observe any signals of zfT1Rs and zfV2Rs probes in solitary cells, while signals of zfT1Rs were observed in the putative taste buds as in the case of PLC-β2, Gia, and G14 (data not shown). As far as we know, there is no other molecular information on SCCs in fish to date. Contrary to SCCs in mammals, these results suggest that SCCs in fish exhibit molecular features distinct from that of taste buds.
Fig. 9. Expression of G14 and Gia in epithelial cells of the body surface in zebrafish.
Single- (A–D, F, and G) and double-label (E and H) in situ hybridization was carried out to examine the expression of G14 and Gia in the epithelial cells of zebrafish. B, D, and G show magnified images of the region with signals in A, C, and F, respectively. The signals of zfPLC-β2 mRNAs were clustered (A and B). The signals of G14 and Gia mRNAs were observed in the clustered zfPLC-β2-expressing cells (E and H). Dark brown matter unrelated to this experiment is indicated by white asterisk. n≥3 (number of sections ≥15) for single-label experiments, n=2 (no. of sections ≥10) for double-label experiments. Scale bars: A, C, and F (in F), 50 μm; B, D, E, G, and H (in H), 20 μm.
Discussion
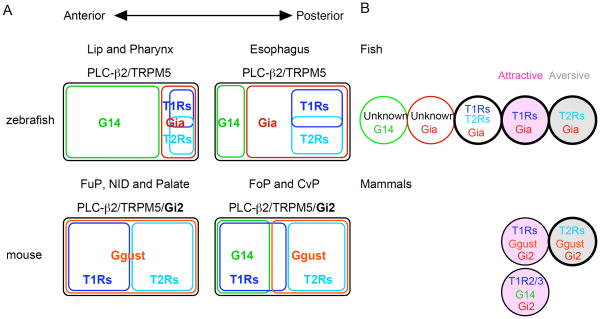
Taste buds are included in the oral and pharyngeal epithelia as gustatory apparatuses in vertebrates. Several traits have been conserved during the evolution of vertebrate gustatory systems including the following: the T1R-mediated attractive behavioral response, the T2R-mediated aversive behavioral response, and the molecular characteristics of T1R- and T2R-expressing TRCs (Chandrashekar et al., 2000; Ishimaru et al., 2005; Li et al., 2002; Mueller et al., 2005; Nelson et al., 2002; Nelson et al., 2001; Oike et al., 2007; Yoshida et al., 2007; Zhang et al., 2003; Zhao et al., 2003). This study adds further molecular information about TRCs expressing PLC-β2 and TRPM5 in zebrafish with regard to their expression of G protein alpha subunits and reveals the unexpected divergence of TRCs in fish (Fig. 10).
Fig. 10. Schematic comparison of PLC-β2/TRPM5-expressing taste receptor cells between zebrafish and mouse.
A, Schematic representation of the expression correlation of G alpha and taste receptor genes and their spatial differences in TRCs expressing both PLC-β2 and TRPM5. Co-expression of Gi2 in these TRCs in mammals is shown in bold above the schemes. B, Schematic representation of the variation in TRCs expressing both PLC-β2 and TRPM5. TRCs are depicted as circles, and the G alpha subunits and taste receptors expressed within them are indicated. T1Rs-and T2Rs-expressing TRCs related to attractive and aversive behaviors are shown in pink and gray, respectively. Bold circles indicate greater variety due to the expression profiles of T1Rs in fish and T2Rs in human. Newly identified subsets of TRCs in fish likely express GPCRs other than the currently known chemosensory receptors, including V2Rs, V1Rs, ORs, TAARs, and ORAs which are expressed in olfactory receptor neurons.
G protein alpha subunits expressed in taste buds: redundancy in mammals and non-redundancy in fish
A comprehensive analysis of the Gna genes specifically expressed in rodent taste buds on the tongue epithelium using differential screening of DNA microarray data and sequential histochemical studies revealed four taste bud-specific Gna genes. Although we were not able to identify the cell type expressing Gq due to its low level of expression, previous studies of the expression profile of Ggust, Gi2, and G14 have indicated redundant expression of Gi2 with Ggust and G14 in PLC-β2/TRPM5-expressing TRCs in rodents (Asano-Miyoshi et al., 2000; Shindo et al., 2008). In fact, functional compensation by Gi2 and G14 could explain the residual responses to sweet, umami, and bitter tastes seen in Ggust knockout mice (Wong et al., 1996). In zebrafish, Gia is expressed in PLC-β2-expressing TRCs, but no other Gq or Gi species were co-expressed in the TRCs, suggesting the involvement of Gia in the taste signaling cascade. Although Gq species are generally upstream of PLC, it has been suggested that heterotrimeric G proteins containing Gi proteins can also activate PLC-β2 through the release of their βγ-subunits (Camps et al., 1992; Kats et al., 1992). In addition, the co-expression of Gia with T1Rs and T2Rs in fish TRCs suggests that Gi species can couple with these class A and C GPCRs. This again highlights the possibility that in mammalian TRCs expressing PLC-β2, Gi2 can mediate taste signals from mammalian T1Rs and T2Rs to PLC-β2 (Kusakabe et al., 2000). Gi2 knockout mice should provide insight into the contributions of Gi species to the taste signaling cascade and the potential roles of Gi2 in cells other than PLC-β2/TRPM5-expressing TRCs.
Molecular evolution of G alpha subunit proteins in taste receptor cells in mammals and fish
An extensive search for Gna genes in fish genomes revealed that teleost genomes contain twice as many genes encoding numerous G protein alpha subunits compared to tetrapod genomes, other than the Gz and Gt families (Fig. 2). This could be explained by a whole-genome duplication event in the most recent common ancestor of teleosts rather than recent gene duplication events (Crollius and Weissenbach, 2005). The fact that the branching of the closest pairs of Gna genes (e.g., G11-1/G11-2, Gq/GqL, and Gi2/Gi2L) in a given fish occurred earlier than the divergence of orthologs among fish species supports the ancestral gene duplication event hypothesis (Robinson-Rechavi et al., 2001). Gia, which is expressed in the PLC-β2-expressing TRCs in fish, is the closest relative to fish Gi1, and thought to be a paralog of zebrafish Gi1. However, after whole genome duplication, extensive gene deletion (i.e., diploidization) occurred, and only a small proportion of paralogous genes were retained as sources of functional innovation (Wolfe, 2001). Retention of paralogs in many G alpha genes suggests close relation of G alpha proteins to the evolution of teleost fish.
We did not identify a Ggust ortholog in the fish or amphibian genomes by our homology-based in silico search. This method was the same as used in identifying Gv, a novel class of G protein alpha subunit (Oka et al., 2009). Therefore, it would be more reasonable to hypothesize that a Ggust ortholog does not exist in the teleost fish genomes rather than to assume that our method failed to find it. This hypothesis suggests that Ggust could have been lost during the evolution of teleost fish after the divergence of lineages between teleost fish and tetrapods. We have no data regarding the expression of G alpha subunits in reptilian or avian TRCs, but Ggust is co-expressed with Gi2 in PLC-β2-expressing TRCs in mammals, despite the fact that Gi2 might be involved in mediating intracellular taste signaling as discussed above. Ggust belongs to the Gt family, which is known to activate phosphodiesterase, so it appears contradictory for Ggust to play a central role in intracellular taste signaling where PLC-β2 is involved (Zhang et al., 2003). Ggust knockout mice, however, exhibit a significant reduction in their responses to sweet, umami, and bitter tastes (Ruiz et al., 2003; Wong et al., 1996). These findings suggest that vertebrates may have retained Ggust, adapted to terrestrial life, and optimized the amplitude of taste signals in TRCs, probably in association with the molecular evolution of T1Rs and T2Rs. Expression of Gia in TRCs in teleost fish might be a new function that teleost fish acquired to compensate for the loss of Ggust during their evolution. In vitro experiments using heterologous expression systems may reveal differences in the physiological characteristics of Gi2 and Ggust, which could provide crucial insights into the molecular and cellular evolution of the peripheral gustatory system in vertebrates.
Diversified taste receptor cells in fish
The total population of T1R- and T2R-expressing cells is much less than that of PLC-β2/TRPM5-expressing TRCs in fish. It remains unclear what the TRCs that do not express T1R or T2R genes actually represent. This study revealed that PLC-β2/TRPM5-expressing TRCs can be divided into two subsets based on their expression of Gia or G14, while known T1R and T2R genes in zebrafish are restricted to a subset of Gia-expressing TRCs. These findings clearly indicate that at least one novel subset of TRCs exists in fish and expresses G14 without expressing any T1R and T2R genes. The TRCs expressing Gia without known T1R or T2R genes may also represent another novel subset in fish (Fig. 6). It will be interesting to identify what kind(s) of receptors, likely GPCRs, are expressed by these TRCs. One possibility is the existence of unidentified T2R genes in Gia-expressing TRCs. The bitter TRCs in humans exhibit a variety of expression profiles of TAS2R (T2R) genes (Behrens et al., 2007), although in rodents, each bitter TRC expresses most T2R genes (Adler et al., 2000). The expression profile of T2R genes identified to date in fish varies like that of humans (Okada et al., 2010), and thus, unidentified T2R genes, if any, would be expressed in a remaining subset of Gia-expressing TRCs without known T1R or T2R genes. However, amino acids received by T1Rs and bitter compounds received by T2Rs are not the entire taste world of aquatic vertebrates (Hara, 1994). Fatty acids, bile salts, and other compounds also activate fish taste systems, but their receptors are not yet identified (Rolen and Caprio, 2008; Yoshii et al., 1979). Some mammalian GPCRs can receive fatty acids (Hirasawa et al., 2005; Itoh et al., 2003), and fish olfactory sensory neurons are activated by bile salts in the fish olfactory system (Døving et al., 1980; Li et al., 1993), although no GPCR belonging to class A or C and known to be involved in chemical sensing (e.g., V1Rs (class A) closely related to T2R, olfactory receptors (class A), TAAR (class A), V2Rs (class C), and ORAs (class A)) is expressed in any taste bud (Fig. 7) (Saraiva and Korsching, 2007). The identification of receptor genes expressed by fish TRCs and the characterization of their ligands are important issues to be resolved in the near future.
Fish genomes contain multiple T1R2 genes, probably the result of a whole-genome duplication event in the most recent common ancestor of teleosts and subsequent gene diversification, as observed for other chemosensory receptor genes (Hashiguchi and Nishida, 2007; Niimura and Nei, 2006) (and references therein), whereas mammalian genomes have a unique T1R2 gene (Ishimaru et al., 2005; Shi and Zhang, 2005). This difference gives rise to a variation to T1R-expressing TRCs in fish compared to mammals (Ishimaru et al., 2005). In addition, fish possess novel subsets of PLC-β2/TRPM5-expressing TRCs, as described above, and seem to have adapted to their environment by diversifying TRCs during their evolution. Taste is a chemical sense closely related to food intake behavior. Insight into how fish have expanded the variation of their TRCs and how mammals have lost this variation would contribute to a better understanding of not only the evolution of vertebrate gustatory systems but also their strategies for survival. A long-term goal is to define the biological importance of the gustatory system in vertebrates.
Supplementary Material
Acknowledgments
Grant information: Grant-in-Aid for Scientific Research to M.O. (21780121), to K.A. (16108004), and to I.M. (18688020) from the Ministry of Education, Culture, Sports, Science and Technology of Japan, and 2006 DIJ Research Grant from the Danone Institute of Japan to I.M.
The authors thank Dr. Y. Sato and Dr. Y. Yoshihara for the plasmids of zfORs, V2Rs, and TAAR9 cDNA and sharing the unpublished information on zfTAAR9.
Literature cited
- Adler E, Hoon MA, Mueller KL, Chandrashekar J, Ryba NJP, Zuker CS. A novel family of mammalian taste receptors. Cell. 2000;100:693–702. doi: 10.1016/s0092-8674(00)80705-9. [DOI] [PubMed] [Google Scholar]
- Asano-Miyoshi M, Abe K, Emori Y. Co-expression of calcium signaling components in vertebrate taste bud cells. Neurosci Lett. 2000;283:61–64. doi: 10.1016/s0304-3940(00)00911-3. [DOI] [PubMed] [Google Scholar]
- Barlow LA, Northcutt RG. Embryonic origin of amphibian taste buds. Dev Biol. 1995;169:273–285. doi: 10.1006/dbio.1995.1143. [DOI] [PubMed] [Google Scholar]
- Behrens M, Foerster S, Staehler F, Raguse J-D, Meyerhof W. Gustatory expression pattern of the human TAS2R bitter receptor gene family reveals a heterogenous population of bitter responsive taste receptor cells. J Neurosci. 2007;27:12630–12640. doi: 10.1523/JNEUROSCI.1168-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camps M, Carozzi A, Schnabel P, Scheer A, Parker PJ, Gierschik P. Isozyme-selective stimulation of phospholipase C-β2 by G protein βγ subunits. Nature. 1992;360:684–686. doi: 10.1038/360684a0. [DOI] [PubMed] [Google Scholar]
- Chandrashekar J, Hoon MA, Ryba NJP, Zuker CS. The receptors and cells for mammalian taste. Nature. 2006;444:288–294. doi: 10.1038/nature05401. [DOI] [PubMed] [Google Scholar]
- Chandrashekar J, Mueller KL, Hoon MA, Adler E, Feng L, Guo W, Zuker CS, Ryba NJP. T2Rs function as bitter taste receptors. Cell. 2000;100:703–711. doi: 10.1016/s0092-8674(00)80706-0. [DOI] [PubMed] [Google Scholar]
- Crollius HR, Weissenbach J. Fish genomics and biology. Genome Res. 2005;15:1675–1682. doi: 10.1101/gr.3735805. [DOI] [PubMed] [Google Scholar]
- Downes GB, Gautam N. The G protein subunit gene families. Genomics. 1999;62:544–552. doi: 10.1006/geno.1999.5992. [DOI] [PubMed] [Google Scholar]
- Døving KB, Selset R, Thommsen G. Olfactory sensitivity to bile salts in salmonid fishes. Acta Physiol Scand. 1980;108:123–131. doi: 10.1111/j.1748-1716.1980.tb06509.x. [DOI] [PubMed] [Google Scholar]
- Finger TE. Evolution of taste and solitary chemoreceptor cell systems. Brain Behav Evol. 1997;50:234–243. doi: 10.1159/000113337. [DOI] [PubMed] [Google Scholar]
- Finger TE, Bryant BP, Kalinoski DL, Teeter JH, Böttger B, Grosvenor W, Cagan RH, Brand JG. Differential localization of putative amino acid receptors in taste buds of the channel catfish, Ictalurus punctatus. J Comp Neurol. 1996;373:129–138. doi: 10.1002/(SICI)1096-9861(19960909)373:1<129::AID-CNE11>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- Finger TE, Böttger B, Hansen A, Anderson KT, Alimohammadi H, Silver WL. Solitary chemoreceptor cells in the nasal cavity serve as sentinels of respiration. Proc Natl Acad Sci USA. 2003;100:8981–8986. doi: 10.1073/pnas.1531172100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara TJ. Olfaction and gustation in fish: an overview. Acta Physiol Scand. 1994;152:207–217. doi: 10.1111/j.1748-1716.1994.tb09800.x. [DOI] [PubMed] [Google Scholar]
- Hashiguchi Y, Nishida M. Evolution of trace amine-associated receptor (TAAR) gene family in vertebrates: lineage-specific expansions and degradations of a second class of vertebrate chemosensory receptors expressed in the olfactory epithelium. Mol Biol Evol. 2007;24:2099–2107. doi: 10.1093/molbev/msm140. [DOI] [PubMed] [Google Scholar]
- Hirasawa A, Tsumaya K, Awaji T, Katsuma S, Adachi T, Yamada M, Sugimoto Y, Miyazaki S, Tsujimoto G. Free fatty acids regulate gut incretin glucagon-like peptide-1 secretion through GPR120. Nat Med. 2005;11:90–94. doi: 10.1038/nm1168. [DOI] [PubMed] [Google Scholar]
- Hoon MA, Adler E, Lindemeier J, Battey JF, Ryba NJP, Zuker CS. Putative mammalian taste receptors: a class of taste-specific GPCRs with distinct topographic selectivity. Cell. 1999;96:541–551. doi: 10.1016/s0092-8674(00)80658-3. [DOI] [PubMed] [Google Scholar]
- Ishimaru Y, Okada S, Naito H, Nagai T, Yasuoka A, Matsumoto I, Abe K. Two families of candidate taste receptors in fishes. Mech Dev. 2005;122:1310–1321. doi: 10.1016/j.mod.2005.07.005. [DOI] [PubMed] [Google Scholar]
- Itoh Y, Kawamata Y, Harada M, Kobayashi M, Fujii R, Fukusumi S, Ogi K, Hosoya M, Tanaka Y, Uejima H, Tanaka H, Maruyama M, Satoh R, Okubo S, Kizawa H, Komatsu H, Matsumura F, Noguchi Y, Shinohara T, Hinuma S, Fujisawa Y, Fujino M. Free fatty acids regulate insulin secretion from pancreatic β cells through GPR40. Nature. 2003;422:173–176. doi: 10.1038/nature01478. [DOI] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma K-i, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kats A, Wu D, Simon MI. Subunits βγ of heterotrimeric G protein activateβ2 isoform of phospholipase C. Nature. 1992;360:686–689. doi: 10.1038/360686a0. [DOI] [PubMed] [Google Scholar]
- Kim M-R, Kusakabe Y, Miura H, Shindo Y, Ninomiya Y, Hino A. Regional expression patterns of taste receptors and gustducin in the mouse tongue. Biochem Biophys Res Commun. 2003;312:500–506. doi: 10.1016/j.bbrc.2003.10.137. [DOI] [PubMed] [Google Scholar]
- Kotrschal K, Krautgartner W-D, Hansen A. Ontogeny of the solitary chemosensory cells in the zebrafish, Danio rerio. Chem Senses. 1997;22:111–118. doi: 10.1093/chemse/22.2.111. [DOI] [PubMed] [Google Scholar]
- Kusakabe Y, Yamaguchi E, Tanemura K, Kameyama K, Chiba N, Arai S, Emori Y, Abe K. Identification of two α-subunit species of GTP-binding proteins, Gα15 and Gαq, expressed in rat taste buds. Biochim Biophys Acta. 1998;1403:265–272. doi: 10.1016/s0167-4889(98)00062-7. [DOI] [PubMed] [Google Scholar]
- Kusakabe Y, Yasuoka A, Asano-Miyoshi M, Iwabuchi K, Matsumoto I, Arai S, Emori Y, Abe K. Comprehensive study on G protein α-subunit in taste bud cells, with special reference to the occurrence of Gαi2 as a major Gα species. Chem Senses. 2000;25:525–531. doi: 10.1093/chemse/25.5.525. [DOI] [PubMed] [Google Scholar]
- Li W, Sorensen PW, Gallaher DD. The olfactory system of migratory adult sea lamprey (Petromyzon marinus) is specifically and acutely sensitive to unique bile salts released by conspecific larvae. J Gen Physiol. 1993;105:569–587. doi: 10.1085/jgp.105.5.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Staszewski L, Xu H, Durick K, Zoller M, Adler E. Human receptors for sweet and umami taste. Proc Natl Acad Sci USA. 2002;99:4692–4696. doi: 10.1073/pnas.072090199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto I, Emori Y, Ninomiya Y, Abe K. A comparative study of three cranial sensory ganglia projecting into the oral cavity: in situ hybridization analyses of neurotrophin receptors and thermosensitive cation channels. Brain Res Mol Brain Res. 2001;93:105–112. doi: 10.1016/s0169-328x(01)00129-2. [DOI] [PubMed] [Google Scholar]
- McLaughlin SK, McKinnon PJ, Margolskee RF. Gustducin is a taste-cell-specific G protein closely related to the transducins. Nature. 1992;357:563–569. doi: 10.1038/357563a0. [DOI] [PubMed] [Google Scholar]
- Mueller KL, Hoon MA, Erlenbach I, Chandrashekar J, Zuker CS, Ryba NJP. The receptors and coding logic for bitter taste. Nature. 2005;434:225–229. doi: 10.1038/nature03352. [DOI] [PubMed] [Google Scholar]
- Nelson G, Chandrashekar J, Hoon MA, Feng L, Zhao G, Ryba NJP, Zuker CS. An amino-acid taste receptor. Nature. 2002;416:199–202. doi: 10.1038/nature726. [DOI] [PubMed] [Google Scholar]
- Nelson G, Hoon MA, Chandrashekar J, Zhang Y, Ryba NJP, Zuker CS. Mammalian sweet taste receptors. Cell. 2001;106:381–390. doi: 10.1016/s0092-8674(01)00451-2. [DOI] [PubMed] [Google Scholar]
- Niimura Y, Nei M. Evolutionary dynamics of olfactory and other chemosensory receptor genes in vertebrates. J Hum Genet. 2006;51:505–517. doi: 10.1007/s10038-006-0391-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohmoto M, Matsumoto I, Misaka T, Abe K. Taste receptor cells express voltage-dependent potassium channels in a cell age-specific manner. Chem Senses. 2006;31:739–746. doi: 10.1093/chemse/bjl016. [DOI] [PubMed] [Google Scholar]
- Ohmoto M, Matsumoto I, Yasuoka A, Yoshihara Y, Abe K. Genetic tracing of the gustatory and trigeminal neural pathways originating from T1R3-expressing taste receptor cells and solitary chemoreceptor cells. Mol Cell Neurosci. 2008;38(4):505–517. doi: 10.1016/j.mcn.2008.04.011. [DOI] [PubMed] [Google Scholar]
- Oike H, Nagai T, Furuyama A, Okada S, Aihara Y, Ishimaru Y, Marui T, Matsumoto I, Misaka T, Abe K. Characterization of ligands for fish taste receptors. J Neurosci. 2007;27(21):5584–5592. doi: 10.1523/JNEUROSCI.0651-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka Y, Saraiva LR, Kwan YY, Korsching SI. The fifth class of Gα proteins. Proc Natl Acad Sci USA. 2009;106:1484–1489. doi: 10.1073/pnas.0809420106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada S, Nakamura S, Nagai T, Ishimaru Y, Matsumoto I, Ieki T, Misaka T, Abe K. Segregated populations of fish taste bud cells express T2R bitter taste receptor genes in a genomic cluster-dependent manner. St. Petersberg, Florida, U.S.A: 2010. [Google Scholar]
- Pfister P, Randall J, Montoya-Burgos JI, Rodriguez I. Divergent evolution among teleost V1r receptor genes. PLoS One. 2007;2(4):e379. doi: 10.1371/journal.pone.0000379. 310.1371/journal.pone.0000379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfister P, Rodriguez I. Olfactory expression of a single and highly variable V1r pheromone receptor-like gene in sigh species. Proc Natl Acad Sci USA. 2005;102:5489–5494. doi: 10.1073/pnas.0402581102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson-Rechavi M, Marchand O, Escriva H, Laudet V. An ancestral whole-genome duplication may not have been responsible for the abundance of duplicated fish genes. Curr Biol. 2001;11:R458–R459. doi: 10.1016/s0960-9822(01)00280-9. [DOI] [PubMed] [Google Scholar]
- Rolen SH, Caprio J. Bile salts are effective taste stimuli in channel catfish. J Exp Biol. 2008;211:2786–2791. doi: 10.1242/jeb.018648. [DOI] [PubMed] [Google Scholar]
- Ruiz CJ, Wray K, Delay ER, Margolskee RF, Kinnamon SC. Behavioral evidence for a role of α-gustducin in glutamate taste. Chem Senses. 2003;28:573–579. doi: 10.1093/chemse/bjg049. [DOI] [PubMed] [Google Scholar]
- Saraiva LR, Korsching SI. A novel olfactory receptor gene family in teleost fish. Genome Res. 2007;17:1448–1457. doi: 10.1101/gr.6553207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y, Miyasaka N, Yoshihara Y. Mutually exclusive glomerular innervation by two distinct types of olfactory sensory neurons revealed in transgenic zebrafish. J Neurosci. 2005;25:4889–4897. doi: 10.1523/JNEUROSCI.0679-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y, Miyasaka N, Yoshihara Y. Hierarchical regulation of odorant receptor gene choice and subsequent axonal projection of olfactory sensory neurons in zebrafish. J Neurosci. 2007;27:1606–1615. doi: 10.1523/JNEUROSCI.4218-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi P, Zhang J. Contrasting modes of evolution between vertebrates sweet/umami receptor genes and bitter receptor genes. Mol Biol Evol. 2005;23:292–300. doi: 10.1093/molbev/msj028. [DOI] [PubMed] [Google Scholar]
- Shindo Y, Miura H, Carninci P, Kawai J, Hayashizaki Y, Ninomiya Y, Hino A, Kanda T, Kusakabe Y. Gα14 is a candidate mediator of sweet/umami signal transduction in the posterior region of the mouse tongue. Biochem Biophys Res Commun. 2008;376:504–508. doi: 10.1016/j.bbrc.2008.09.035. [DOI] [PubMed] [Google Scholar]
- Stone LM, Finger TE, Tam PPL, Tan S-S. Taste receptor cells arise from local epithelium, not neurogenic ectoderm. Proc Natl Acad Sci USA. 1995;92:1916–1920. doi: 10.1073/pnas.92.6.1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Tizzano M, Dvoryanchikov G, Barrows JK, Kim S, Chaudhali N, Finger TE. Expression of Galpha14 in sweet-transducing taste receptor cells of the posterior tongue. BMC Neurosci. 2008;9:110. doi: 10.1186/1471-2202-9-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wettschureck N, Offermanns S. Mammalian G proteins and their cell type specific functions. Physiol Rev. 2005;85:1159–1204. doi: 10.1152/physrev.00003.2005. [DOI] [PubMed] [Google Scholar]
- Wolfe KH. Yesterday’s polyploids and the mystery of diploidization. Nat Rev Genet. 2001;2:333–341. doi: 10.1038/35072009. [DOI] [PubMed] [Google Scholar]
- Wong GT, Gannon KS, Margolskee RF. Transduction of bitter and sweet taste by gustducin. Nature. 1996;381:796–800. doi: 10.1038/381796a0. [DOI] [PubMed] [Google Scholar]
- Yasuoka A, Aihara Y, Matsumoto I, Abe K. Phospholipase C-beta 2 as a mammalian taste signaling marker is expressed in the multiple gustatory tissues of medaka fish, Oryzias latipes. Mech Dev. 2004;121:985–989. doi: 10.1016/j.mod.2004.03.009. [DOI] [PubMed] [Google Scholar]
- Yoshida Y, Saitoh K, Aihara Y, Okada S, Misaka T, Abe K. Transient receptor potential channel M5 and phospholipase C-beta2 colocalizing in zebrafish taste receptor cells. Neuroreport. 2007;18:1517–1520. doi: 10.1097/WNR.0b013e3282ec6874. [DOI] [PubMed] [Google Scholar]
- Yoshii K, Kamo N, Kurihara K, Kobatake Y. Gustatory responses of eel palatine receptors to amino acids and carboxylic acids. J Gen Physiol. 1979;74:301–317. doi: 10.1085/jgp.74.3.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Hoon MA, Chandrashekar J, Mueller KL, Cook B, Wu D, Zuker CS, Ryba NJP. Coding of sweet, bitter, and umami tastes: different receptor cells sharing similar signaling pathways. Cell. 2003;112:293–301. doi: 10.1016/s0092-8674(03)00071-0. [DOI] [PubMed] [Google Scholar]
- Zhao GQ, Zhang Y, Hoon MA, Chandrashekar J, Erlenbach I, Ryba NJP, Zuker CS. The receptors for mammalian sweet and umami taste. Cell. 2003;115:255–266. doi: 10.1016/s0092-8674(03)00844-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.