Abstract
Saliva of blood-sucking arthropods contains a complex mixture of peptides that affect their host’s hemostasis, inflammation, and immunity. These activities can also modify the site of pathogen delivery and increase disease transmission. Saliva also induces hosts to mount an antisaliva immune response that can lead to skin allergies or even anaphylaxis. Accordingly, knowledge of the salivary repertoire, or sialome, of a mosquito is useful to provide a knowledge platform to mine for novel pharmacological activities, to develop novel vaccine targets for vector-borne diseases, and to develop epidemiological markers of vector exposure and candidate desensitization vaccines. The mosquito Ochlerotatus triseriatus is a vector of La Crosse virus and produces allergy in humans. In this work, a total of 1,575 clones randomly selected from an adult female O. triseriatus salivary gland cDNA library was sequenced and used to assemble a database that yielded 731 clusters of related sequences, 560 of which were singletons. Primer extension experiments were performed in selected clones to further extend sequence coverage, allowing for the identification of 159 protein sequences, 66 of which code for putative secreted proteins. Supplemental spreadsheets containing these data are available at http://exon.niaid.nih.gov/transcriptome/Ochlerotatus_triseriatus/S1/Ot-S1.xls and http://exon.niaid.nih.gov/transcriptome/Ochlerotatus_triseriatus/S2/Ot-S2.xls.
Keywords: mosquito, salivary gland, sialome, transcriptome, La Crosse virus
When attempting to blood feed on a mammalian host, mosquitoes face their hosts’ mechanisms to prevent blood loss, the hemostasis system (Ribeiro and Arca 2009), as well as their host-defensive behaviors (Edman and Kale 1971, Edman et al. 1974) that are triggered by pain and itching at the site of the bite. Mammalian hemostasis relies on a complex set of reactions, including platelet aggregation, blood clotting, and vasoconstriction, whereas inflammatory reactions caused by tissue injury or immune reactions lead to pain, itching, and edema. A few species of mosquitoes studied to date indicate that their saliva contains antiplatelet, vasodilatory, anti-clotting, and immunomodulators that might help blood feeding (Ribeiro and Arca 2009). Indeed, mosquitoes deprived of salivary function have difficulty initiating a blood meal (Mellink and Van Den Boven Kamp 1981, Rossignol et al. 1984).
In addition to its role in blood feeding, mosquito saliva also plays an important biological function in sugar feeding, as well as being a vehicle for pathogen delivery. Antimicrobial peptides are also present in male and female mosquito saliva, possibly checking the growth of microbes contaminating the sugar meal. Possibly because of the pharmacologically active components of mosquito saliva, saliva can also enhance pathogen transmission when compared with needle injection, and may cause allergic skin reactions in their hosts (Ribeiro and Francischetti 2003, Ribeiro and Arca 2009).
The mosquito Ochlerotatus triseriatus (formerly known as Aedes triseriatus) (Reinert 2000) is a common tree hole mosquito found east of the Rocky Mountains in the United States, and an important vector of La Crosse virus (Craig 1983, DeFoliart 1983), the agent of a zoonosis causing human encephalitis in the United States (Rust et al. 1999, Romero and Newland 2003). O. triseriatus saliva is known to produce allergic responses (Peng et al. 1998), and is implicated in potentiating virus transmission (Edwards et al. 1998). Salivary apyrase, which breaks down adenosine 5′-diphosphate and adenosine triphosphate (agonists of platelet and neutrophil aggregation) (Reno and Novak 2005), and vasodilatory (Ribeiro et al. 1994) activities have been reported in O. triseriatus, but no salivary protein has been molecularly characterized to date. In this study, we describe the sialotranscriptome of O. triseriatus to help understand the evolution of blood feeding in mosquitoes, to provide a knowledge platform for functional studies, and to provide targets for vaccine and epidemiological markers of vector exposure.
Materials and Methods
Mosquitoes and cDNA Library Construction
Mosquitoes used in this work are from a colony originated from field material collected near La Crosse, Wisconsin, in 1981, and have been colonized continuously at the Colorado State University Arthropod-Borne and Infectious Diseases Laboratory (Fort Collins, CO), at 70°F, 70% RH, 16-h light, 8-h dark cycles. Because messenger RNA, but not ribosomal RNA, is polyadenylated (PolyA), PolyA containing RNA was extracted from 80 dissected pairs of salivary glands using the Micro-FastTrack mRNA isolation kit (Invitrogen, Carlsbad, CA), which was then used to make a polymerase chain reaction (PCR)-based cDNA library using the SMART cDNA library construction kit (BD Biosciences-Clontech, Palo Alto, CA), as described before (Valenzuela et al. 2002a).
CDNA Sequencing
The salivary gland cDNA library was plated on Luria-Bertani/MgSO4 plates containing 5-bromo-4-chloro-3-indolyl β-D-galactoside/isopropyl β-D-thiogalactoside to an average of 250 plaques per 150-mm petri plate. Recombinant (white) plaques were randomly selected and transferred to 96-well MICROTEST TM U-bottom plates (BD Biosciences, Franklin Lakes, NJ), containing 100 μl of SM buffer (0.1 M NaCl, 0.01 M MgSO4, 7 H2O, 0.035 M Tris HCl (pH 7.5), and 0.01% gelatin) per well. The plates were covered and placed on a gyrating shaker for 30 min at room temperature. The phage suspension was either immediately used for PCR or stored at 4°C for future use.
To amplify the cDNA using a PCR reaction, 4 μl of the phage sample was used as a template. The primers were sequences from the λ TriplEx2 vector and named pTEx2 5seq (5′-TCC GAG ATC TGG ACG AGC-3′) and pTEx2 3LD (5′-ATA CGA CTC ACT ATA GGG CGA ATT GGC-3′), positioned at the 5′ end and the 3′ end of the cDNA insert, respectively, as done in our previous studies (Valenzuela et al. 2002a). The reaction was carried out in 96-well flexible PCR plates (Fisher Scientific, Pittsburgh, PA) using TaKaRa EX Taq polymerase (Takara Mirus Bio, Madison, WI) on a GeneAmp PCR system 9700 (PerkinElmer, Foster City, CA). The PCR conditions were as follows: one hold of 95°C for 3 min; 25 cycles of 95°C for 1 min; 61°C for 30 s; and 72°C for 5 min. The amplified products were analyzed on a 1.5% agarose/EtBr gel. cDNA library clones were PCR amplified, and the ones showing single band were selected for sequencing. Approximately 200–250 ng of each PCR product was transferred to Thermo Fast 96-well PCR plates (ABgene, Epsom, Surray, United Kingdom) and frozen at −20°C. Samples were shipped on dry ice to the Rocky Mountain Laboratories Genomics Unit with primer and template combined together in an ABI 96-well Optical Reaction Plate (P/N 4306737) following the manufacturer’s recommended concentrations. Sequencing reactions were set up as recommended by Applied Biosystems BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA) by adding 1 μl of ABI BigDye Terminator Ready Reaction Mix v3.1 (P/N 4336921), 3 μl of 5× ABI Sequencing Buffer (P/N 4336699), and 2 μl of water for a final volume of 10 μl. Cycle sequencing was performed at 96°C for 10 s, 50°C for 5 s, and 60°C for 4 min for 27 cycles on either a Bio-Rad Tetrad 2 (Bio-Rad Laboratories, Hercules, CA) or ABI 9700 (Applied Biosystems, Foster City, CA) thermal cycler. Fluorescently labeled extension products were purified following Applied Biosystems BigDye XTerminator Purification protocol and subsequently processed on an ABI 3730xL DNA Analyzer (Applied Biosystems, Foster City, CA). The AB1 file generated for each sample from the 3730xL DNA Analyzer was provided to researchers in Rockville, Maryland, through a secure network drive for all subsequent downstream sequencing analysis. In addition to the sequencing of the cDNA clones, primer extension experiments were performed in selected clones to further extend sequence coverage.
Bioinformatic Tools and Procedures
Expressed sequence tags (EST) were trimmed of primer and vector sequences. The BLAST tool (Altschul et al. 1997), CAP3 assembler (Huang and Madan 1999), and ClustalW (Thompson et al. 1997) software were used to compare, assemble, and align sequences, respectively. Phylogenetic analysis and statistical neighbor-joining (NJ) bootstrap tests of the phylogenies were done with the Mega package (Kumar et al. 2004). For functional annotation of the transcripts, we used the tool BlastX (Altschul et al. 1997) to compare the nucleotide sequences with the nonredundant (NR) protein database of the National Center for Biotechnology Information (NCBI; National Library of Medicine, National Institutes of Health, Bethesda, MD) and with the Gene Ontology database (Ashburner et al. 2000). The tool, reverse position-specific Blast (RPSBLAST) (Altschul et al. 1997), was used to search for conserved protein domains in the Pfam (Bateman et al. 2000), SMART (Schultz et al. 2000), Kog (Tatusov et al. 2003), and conserved domains databases (Wheeler et al. 2005). We have also compared the transcripts with other subsets of mitochondrial and rRNA nucleotide sequences downloaded from NCBI and with several organism proteomes downloaded from NCBI, ENSEMBL, or VectorBase. Segments of the three-frame translations of the EST (because the libraries were unidirectional, six-frame translations were not used), starting with a methionine found in the first 300 predicted amino acids, or the predicted protein translation in the case of complete coding sequences, were submitted to the SignalP server (Nielsen et al. 1997) to help identify translation products that could be secreted. O-glycosylation sites on the proteins were predicted with the program NetOGlyc (Julenius et al. 2005). Functional annotation of the transcripts was based on all the comparisons above. After inspection of all these results, transcripts were classified as either secretory (S), housekeeping (H), or of unknown (U) function, with further subdivisions based on function and/or protein families. Codon volatility was calculated, as previously described (Friedman and Hughes 2005).
Results
Characteristics of the Assembled Salivary EST Set
A total of 1,575 cDNA clones was used to assemble a database (Supplemental Table S1) that yielded 731 clusters of related sequences, including 560 singletons. The 731 clusters were compared, using the programs BlastX, BlastN, or RPSBLAST (Altschul et al. 1997), with the NR protein database of the NCBI, with the gene ontology database (Ashburner et al. 2000), with the conserved domains database of the NCBI (Marchler-Bauer et al. 2002), and with a custom-prepared subset of the NCBI nucleotide database containing either mitochondrial or rRNA sequences.
Manual annotation of the contigs indicated that 39% of the ESTs assembled in 182 contigs attributed to messages coding for putative secreted proteins (Table 1). This category also had the highest ratio of ESTs per contig, namely 3.5. Twenty-two percent of the ESTs could not be classified and belonged to the unknown category. These can represent novel proteins or derive from the less conserved 3′ or 5′ untranslated regions of genes, as was indicated for the sialotranscriptome of Anopheles gambiae (Arca et al. 2005). Transcripts associated to the protein synthetic machinery accounted for 20% of the transcriptome, containing an average of 2.6 ESTs per contig. The remaining transcripts code for other H functions, as indicated in Table 1, and have <2 ESTs per contig. This distribution is typical of previous mosquito sialotranscriptomes (Valenzuela et al. 2003; Ribeiro et al. 2004, 2007; Arca et al. 2007; Calvo et al. 2007a).
Table 1.
Functional classification of the transcriptome and their abundance
| Class | No. contigs | No. ESTs | ESTs/Contig | % total ESTs |
|---|---|---|---|---|
| Secreted | 182 | 645 | 3.5 | 39.0 |
| Unknown | 244 | 355 | 1.5 | 21.5 |
| Protein synthesis machinery | 127 | 328 | 2.6 | 19.9 |
| Unknown, conserved | 73 | 86 | 1.2 | 5.2 |
| Metabolism, energy | 38 | 74 | 1.9 | 4.5 |
| Transcription machinery | 13 | 24 | 1.8 | 1.5 |
| Signal transduction | 11 | 15 | 1.4 | 0.9 |
| Protein export machinery | 8 | 12 | 1.5 | 0.7 |
| Proteasome machinery | 7 | 7 | 1.0 | 0.4 |
| Transporters/storage | 6 | 6 | 1.0 | 0.4 |
| Protein modification machinery | 4 | 4 | 1.0 | 0.2 |
| Metabolism, carbohydrate | 4 | 4 | 1.0 | 0.2 |
| Metabolism, lipid | 3 | 3 | 1.0 | 0.2 |
| Cytoskeletal | 2 | 3 | 1.5 | 0.2 |
| Nuclear regulation | 2 | 2 | 1.0 | 0.1 |
| Transcription factor | 2 | 2 | 1.0 | 0.1 |
| Metabolism, nucleotide | 2 | 2 | 1.0 | 0.1 |
| Metabolism, amino acid | 2 | 2 | 1.0 | 0.1 |
| Oxidant metabolism/detoxication | 1 | 1 | 1.0 | 0.1 |
| Total | 731 | 1,575 |
Possibly Secreted (S) Class of Expressed Genes
A total of 645 ESTs represents putative O. triseriatus salivary components (Table 2; Supplemental File S1). These include ubiquitous known gene families, e.g., families seen in other organs and organisms, such as enzymes coding for carbohydrate hydrolysis, contributing to over 15% of the ESTs on the secreted class, as well as protein families unique to hematophagous Diptera, or to Culicines alone. Many of these putatively secreted protein families are multigenic, such as the D7/OBP-like families contributing to 11% of all transcripts associated with secreted products, which is less than previously seen in Anopheles or Aedes sialotranscriptomes, probably because of the dilution effect after abundant expression of the polyhistidine/8.3-kDa family and the 9-kda WW family, which together accounts for over 32% of the ESTs of the secreted class.
Table 2.
Classification and abundance of the salivary secretome of Ochlerotatus triseriatus
| Class Subclass | No. ESTs | % |
|---|---|---|
| Enzymes | ||
| Amylase/maltase | 99 | 15.3 |
| Chitinase | 1 | 0.2 |
| Serine proteases | 11 | 1.7 |
| Endonuclease | 1 | 0.2 |
| Protease inhibitor motifs | ||
| Kazal domain | 6 | 0.9 |
| Serpin | 4 | 0.6 |
| Trypsin inhibitor-like domain | 3 | 0.5 |
| Antigen 5/CRISP family | 6 | 0.9 |
| Immunity-related proteins | ||
| Pattern recognition proteins | 23 | 3.6 |
| Antimicrobial peptides | 18 | 2.8 |
| Mucin | 1 | 0.2 |
| Proteins found in Nematocera sialomes | ||
| D7 protein family | 68 | 10.5 |
| Aegyptin/30-kDa antigen | 50 | 7.8 |
| 56-kDa protein family | 28 | 4.3 |
| Glycine-rich family: low complexity | 26 | 4.0 |
| 41-kDa family | 2 | 0.3 |
| SGS family | 2 | 0.3 |
| Proline-rich salivary protein | 1 | 0.2 |
| Proteins found in Culicine sialomes | ||
| PolyHis/8.3-kDa family | 144 | 22.3 |
| 9-kDa WW protein family | 65 | 10.1 |
| 34-kDa protein family | 27 | 4.2 |
| 30.5-kDa protein family | 26 | 4.0 |
| Basic tail protein family | 17 | 2.6 |
| 11.6-kDa protein family | 5 | 0.8 |
| 7.8-kDa protein family | 4 | 0.6 |
| Other possible secreted peptides | 7 | 1.1 |
| Total | 645 | 100 |
The Salivary Secretome of O. triseriatus
From the sequenced cDNAs, a total of 159 novel O. triseriatus protein sequences was derived, 66 of which code for putative secreted products (Supplemental File S2). Several of these deducted coding sequences are truncated at their 5′ location, but allow comparison with other sialomes and positively identify members of different salivary protein families within the O. triseriatus sialome.
Proteins Belonging to Ubiquitous Protein Families
Enzymes
Sequences coding for amylases, maltase, chitinase, serine proteases, and a DNase were obtained. Amylase and maltase are a common finding in the sialotranscriptomes of Nematocera, and certainly have a function in sugar feeding. The chitinase finding is not that common, and may be associated with antimicrobial immunity, and indeed there are chitinases of An. gambiae that were found to be overexpressed in bacterial-challenged mosquitoes, such as the protein gi 46095203 (Shi and Paskewitz 2004). The serine proteases cannot be easily functionally characterized, as they may play an endogenous role as activators of immunity, such as prophenoloxidase activators, or in specific roles in blood feeding, such as plasminogen activator or fibrinogenases, as was found for salivary proteases of tabanids (Xu et al. 2008, Ma et al. 2009). The DNase enzyme from O. triseriatus has a clear signal peptide indicative of secretion. With regard to mosquito sialomes, this type of enzyme was previously found solely in Culex quinquefasciatus, and the recombinant enzyme characterized (Calvo and Ribeiro 2006). It is to be noted that transcripts coding for members of the 5′-nucleotidase family, associated with salivary apyrase in mosquitoes (Champagne et al. 1995, Sun et al. 2006), have not been identified in O. triseriatus, although it was abundantly expressed in Aedes aegypti and Aedes albopictus transcriptomes, and the nucleotidase activity previously reported for O. triseriatus salivary gland homogenates was similar to that of Ae. aegypti in amounts and kinetic characterisitics (pH and divalent cation dependence) (Reno and Novak 2005). This may be the result of technical reasons, including the presence of SFI restriction site within the transcript.
Antiprotease Domains
The anticlotting of Ae. aegypti has been previously characterized to be a member of the Serpin family of serine proteinase inhibitors that potently inhibits factor Xa of the clotting cascade (Stark and James 1998). The sialotranscriptome of Ae. aegypti later revealed a second member of this family, still not functionally characterized (Ribeiro et al. 2007). Supplemental Table S2 displays two such proteins from O. triseriatus, OT-639 having top similarity with Aedes factor Xa anticoagulant. Additionally, two peptides contain the Kazal domain, and OT-630 has a typical Trypsin inhibitor-like domain. These peptide families were also found in Aedes and Culex sialotranscriptomes.
Antigen-5 Family
A truncated member of the antigen-5 family is presented in Supplemental Table S2. Members of this family are ubiquitous in nature (Gibbs et al. 2008), in which they have been attributed several functions, including toxic components of snake venom (Yamazaki et al. 2003), proteolytic activity in snails (Milne et al. 2003), and immunoglobulin binding from the salivary glands of the stable fly, Stomoxys calcitrans (Ameri et al. 2008). No functional activity has been reported to date for any mosquito protein of this family.
Immunity-Related Peptides
Sequences for a Gram-negative binding protein, a ficolin, and two C-type lectins are presented in Supplemental Table S2. Three are truncated, except for a C-type lectin. These possibly represent proximal members of immunity cascades involving microbial recognition and activation of proteolytic cascades. Additionally, a homologue of the antimicrobial peptide gambicin (Vizioli et al. 2001) and a homologue of a virus-induced protein from Ae. aegypti, which appears to be a mucin with 12 potential galactosylation sites, are revealed.
Proteins Common to Sialomes of Blood-Sucking Nematocera or Culicidae
D7 Family
The D7 proteins belong to the superfamily of odorant binding proteins (OBP) (Hekmat-Scafe et al. 2000). Mosquito genomes and transcriptomes (Arca et al. 2005, Ribeiro et al. 2007) revealed the multifamily nature of this family, consisting of multiple copies of long (two OBP domains) and short (one OBP domain) versions of the protein. Phlebotomine sand flies also have D7 members expressed in their salivary glands (Valenzuela et al. 2002b). The crystal structures of a short D7 protein from An. gambiae and a long D7 protein from Ae. aegypti revealed that the D7 OBP domains have seven α helices, two more than the canonical OBP family (Mans et al. 2007). Short versions of anopheline D7 mosquitoes were shown to bind biogenic amines such as serotonin and histamine (Calvo et al. 2006). More recently, the amino-terminal OBP domain of a D7 long form of Ae. aegypti was shown to bind peptidic leukotrienes with high affinity (Calvo et al. 2009a). In addition to these inflammatory agonist-binding functions, a short D7 protein from Anopheles stephensi, named hamadarin, was shown to inhibit bradykinin formation by inhibiting the FXII/kallikrein pathway (Isawa et al. 2002). The sialotranscriptome of O. triseriatus revealed four members of the family, two of which appear to be alleles (OT-58 and OT-53).
The 30-kDa Antigen/GE-Rich/Aegyptin Family
The sialotranscriptome of Culex tarsalis allowed the identification of five protein sequences from the 30-kDa antigen/Aegyptin family (Supplemental Table S2), all represented by 1–14 ESTs found in the library. These five sequences are consistent with the existence of two genes providing for alleles and splice variants. The multigene pattern has been observed previously for Aedes and Culex mosquitoes (Ribeiro et al. 2004, 2007; Arca et al. 2007), whereas anophelines to date appear to have only one gene (Valenzuela et al. 2003, Arca et al. 2005, Calvo et al. 2009b). One of the Aedes and one of the Anopheles proteins have been characterized as inhibitors of collagen-induced platelet aggregation by their strong interaction with collagen (Calvo et al. 2007b, Yoshida et al. 2008).
The 41- and 56-kDa Families
These two protein families are uniquely found in sialotranscriptomes of blood-sucking Nematocera (41 kDa) or blood-sucking mosquitoes (56 kDa), and one member of each is described for the sialotranscriptome of O. triseriatus. Their function is unknown to date. Members of these protein families have been shown to be expressed in male and female mosquitoes (Arca et al. 2007, Ribeiro et al. 2007), suggesting a role in sugar feeding rather than blood feeding. Psi-blast search of members of the 56-kDa family against the NR database only retrieved mosquito and bacterial proteins. This fact, plus the single exon structure of these genes in Ae. aegypti and An. gambiae, led to the hypothesis that the 56-kDa protein gene was acquired by horizontal transfer from a bacteria early in mosquito evolution (Arca et al. 2007).
The Basic Tail Family
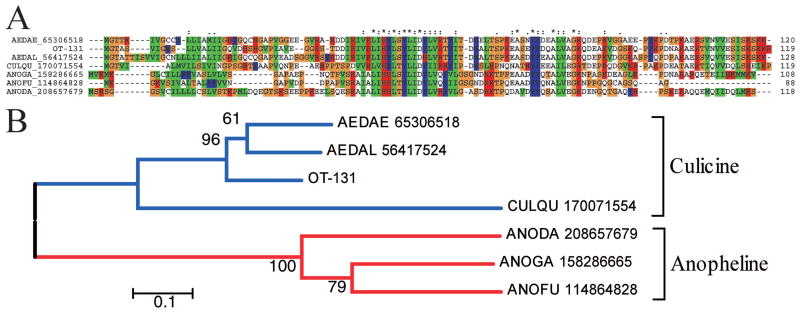
The basic tail proteins are so named because of a cluster of lysine/arginine residues found in the Aedes homologues, and have been found in most mosquito sialotranscriptomes done to date. They present similarities among themselves, but not to other known proteins (Fig. 1A). The bootstrapped phylogram derived from the alignment shows distinct anopheline and culicine clades, as expected from the phylogeny. Transcripts coding for this protein are well represented in the O. triseriatus sialotranscriptome, accounting for 14 ESTs coding for six variants, suggestive of polymorphism.
Fig. 1.
The basic tail family of mosquito salivary proteins. (A) Clustal alignment. The O. triseriatus protein is named OT-131. The remaining sequences are named with the first three letters from the genus name, followed by two letters from the species name and by their NCBI protein accession number. The symbols above the alignment indicate the following: (*) identical sites; (:) conserved sites; (.) less conserved sites. (B) NJ bootstrapped phylogram of the alignment in (A), showing culicine and anopheline clades. The numbers on the branches represent the percentage of bootstrap support. The bar on the bottom represents 10% amino acid divergence. For more details, see text. (Online figure in color.)
Glycine-Rich Polypeptides
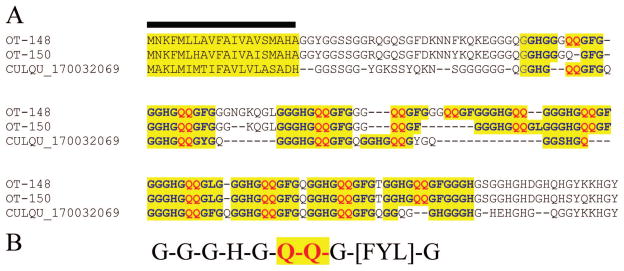
Two kinds of glycine-rich polypeptides can be identified in the O. triseriatus sialotranscriptome. The first kind has unique repeats consisting of the decapeptide unit G-G-G-H-G-Q-Q-G-[FYL]-G perfectly repeated (with different spacing) seven times in OT-48 (Fig. 2A). OT-150 appears to be a splice variant or a product of a recent or conserved gene duplication. A homologous protein was found in the Cx. quinquefasciatus proteome, but not in a previous sialotranscriptome. This Culex protein retains the repeat units, as well as many other identical amino acids. Interestingly, this protein was not found previously in any other sialotranscriptome, despite being relatively well represented in O. triseriatus with nine ESTs. Search of the motif using the tool seedtop against the proteomes of An. gambiae and Ae. aegypti did not reveal the block pattern indicated in Fig. 2B in any of the annotated proteomes. This novel family is named GGHGQQ for its repeat.
Fig. 2.
Members of the GGHGQQ family of culicines. (A) Clustal alignment of two O. triseriatus proteins with a hypothetical protein from Cx. quinquefasciatus. The signal sequence indicative of secretion is marked with a bar; remaining markings refer to repeat structure, indicated in (B). (Online figure in color.)
Search of homologues of OT-48 by the use of the tool blastp against the NR protein database retrieves many matches of other glycine- and histidine-rich proteins of similar size from hymenoptera and Drosophila, including some with closely related repeats such as G-G-G-H-G-X-Q-G, suggesting this protein family may derive from a widespread insect superfamily, possibly related with antimicrobial activity.
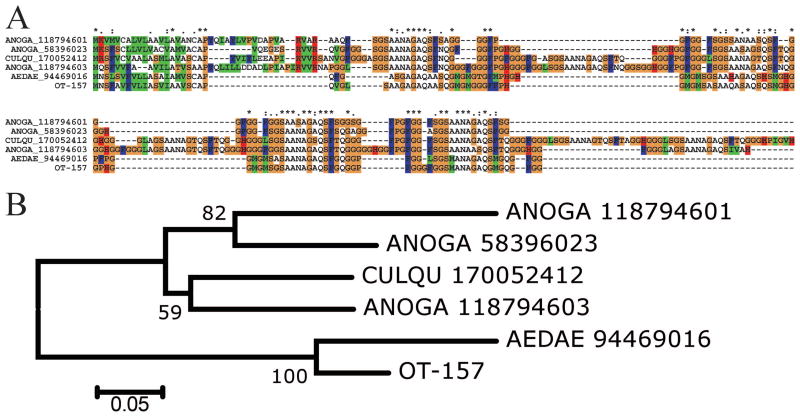
The second glycine-rich peptide was deducted from 11 ESTs and is similar to a protein found in the Ae. aegypti sialotranscriptome (Ribeiro et al. 2007), and to hypothetical proteins found in the genomes of Cx. quinquefasciatus and An. gambiae (Fig. 3). The phylogram indicates this protein diverged considerably from a common ancestor, with at least three genes represented in An. gambiae. As expected, the O. triseriatus sequence strongly clusters with the Ae. aegypti homologue.
Fig. 3.
The Aedes glycine-rich family of proteins. (A) Clustal alignment. The O. triseriatus protein is represented by OT-157. The remaining sequences are named with the first three letters from the genus name, followed by two letters from the species name and by their NCBI protein accession number. The symbols above the alignment indicate the following: (*) identical sites; (:) conserved sites; (.) less conserved sites. (B) NJ bootstrapped phylogram of the alignment in A. The numbers on the branches represent the percentage of bootstrap support. The bar on the bottom represents 5% amino acid divergence. For more details, see text. (Online figure in color.)
The 13-kDa Family
This family of salivary proteins has been previously characterized in the sialome of Ae. albopictus (Arca et al. 2007), in which it was pointed out that homologue proteins were previously found in the sialotranscriptomes of Ae. aegypti, Cx. quinquefasciatus, and male An. gambiae, and have a common framework of 10 cysteines. OT-212 is 65% identical to one of the Ae. albopictus proteins that characterized this family. The function of this protein family is unknown, but may be related to antimicrobial function, as it was found in male mosquito sialotranscriptomes.
The 6.2-kDa Family
Previously, an orphan putative secreted peptide was described in the sialome of Ae. aegypti named putative 8.5-kDa secreted salivary peptide. OT-547 codes for a peptide that is 57% identical to this Aedes protein, and weaker (<40%) identities to other proteins. Interestingly, OT-547 contains the block I-G-x(10)-G-x(2)-W-x(2)-G-x(5)-E-x(2)-R-R-x-F, which was deducted from alignment of anopheline proteins of the 6.2-kDa family described in previous sialotranscriptomes (Arca et al. 2005, Calvo et al. 2007a, 2009b). Alignment of OT-547 to the Aedes and anopheline sequences (Fig. 4) reveals a region of higher amino acid conservation at the carboxy termini, and shows that the anopheline sequences are smaller on their amino-terminal side. These peptides possibly arose from a common mosquito gene that evolved fast. The function of this protein family is unknown, but Ae. aegypti transcripts for this protein were also found in male mosquitoes and female salivary glands, but not in female carcass devoid of salivary glands (Ribeiro et al. 2007), suggesting a role in sugar feeding or as an antimicrobial.
Fig. 4.
The 6.2-kDa family of mosquito salivary peptides. Clustal alignment of the mature versions (signal peptide removed) of the O. triseriatus protein OT-547 with those of Ae. aegypti, An. gambiae, Anopheles funestus, and Anopheles darlingi. These sequences are named with the first three letters from the genus name, followed by two letters from the species name and by their NCBI protein accession number. The symbols below the alignment indicate the following: (*) identical sites; (:) conserved sites; (.) less conserved sites. (Online figure in color.)
The SG3/WW Family
Two polypeptides containing one WW and one WWW motif plus several other W residues have been previously described in Ae. aegypti (Ribeiro et al. 2007). OT-175 has 46% identity to one of these proteins, including the rare WW doublet and one WWF instead of a WWW. These proteins have the block I-F-x(3,8)-W-x(17,26)-P-x(0,8)-W-W-x-R, which was deducted from the alignment from members of the SG3 protein family of anophelines, which was first identified in 1999 (Arca et al. 1999). Alignment of the aedine proteins with those of An. gambiae and An. stephensi suggests that the aedine proteins are shorter versions of the anopheline proteins, with conservation of the two tryptophane clusters (Fig. 5). Transcripts for this family were found expressed in Ae. aegypti female salivary glands and in male mosquitoes, suggesting a role in sugar feeding or as antimicrobial (Ribeiro et al. 2007). The anopheline proteins have many Thr and Ser residues in their carboxy termini that are potentially glycosylated (Julenius et al. 2005), and may thus be characterized as mucins. However, the aedine proteins have only two potential galactosylation sites (Fig. 6).
Fig. 5.
The SG3/WWW family of mosquito salivary peptides. Clustal alignment of the O. triseriatus protein OT-175 with those of Ae. aegypti, An. gambiae, and An. stephensis. These sequences are named with the first three letters from the genus name, followed by two letters from the species name and by their NCBI protein accession number. The symbols below the alignment indicate the following: (*) identical sites; (:) conserved sites; (.) less conserved sites. (Online figure in color.)
Fig. 6.
The WPCWW family of mosquito salivary peptides. Clustal alignment of the mature versions (signal peptide removed) of the O. triseriatus protein OT-86 with those of Ae. aegypti, Ae. albopictus, and Cx. quinquefasciatus. These sequences are named with the first three letters from the genus name, followed by two letters from the species name and by their NCBI protein accession number. The symbols below the alignment indicate the following: (*) identical sites; (:) conserved sites; (.) less conserved sites. The cysteine residues are in black background. (Online figure in color.)
The SGS Family
The SGS family of mosquito proteins was characterized as membrane-bound salivary proteins that were candidate receptors for Plasmodium invasion (Korochkina et al. 2006). Unexpectedly, peptide fragments of members of this family were also found by mass spectrometry in proteomic analysis of Ae. aegypti and An. gambiae saliva (not salivary gland homogenates) (Orlandi-Pradines et al. 2007), suggesting a noncanonical mode of secretion, possibly after proteolysis of a membrane-bound precursor. This protein family is similar to bacterial proteins and may have been acquired early in the evolution of mosquitoes by horizontal transfer (Arca et al. 2005, Korochkina et al. 2006).
Proteins Found to Date Only in Culicines (Not Found in Anophelines)
The 34-kDa Family
Four genes coding for this protein family were identified in the Ae. aegypti sialome, one of which codes for a shorter version of the protein (Ribeiro et al. 2007). Members of this family were also found in Ae. albopictus (Arca et al. 2007) and Cx. quinquefasciatus (Ribeiro et al. 2004) sialomes. OT-110 represents a member of this family from O. triseriatus, having 61% sequence identity with the Ae. albopictus homologue. The function of this protein family is unknown.
The 30.5-kDa Family
This family was previously characterized in Ae. aegypti, with members also found in the sialomes of Ae. albopictus, Cx. quinquefasciatus, and Cx. tarsalis. Related sequences were not found outside of the culicines. Psi-blast of members of this protein family against the NR protein databases converged after six iterations, pulling together solely mosquito proteins. OT-114, OT-118, and OT-117 represent possibly alleles of the same gene found expressed in the sialotranscriptome of O. triseriatus. The function of this protein family is unknown.
The WPCWW Family
Previous sialotranscriptomes identified several proteins by the generic name of W or tryptophane rich. OT-84, OT-86, OT-87, and OT-89 code for slightly different (possibly alleles) members of the W-rich family, previously described in Aedes and Culex (but not anopheline) sialotranscriptomes (Ribeiro et al. 2004, 2007; Arca et al. 2007). Alignment of these sequences (Fig. 6) shows a conserved framework, in particular a block with three tryptophanes, W-P-x-W-x-G-L-G-P-x-I-P-P-x-W-x-R, located at the first half of the sequence. Notice also that the aedine proteins contain two cysteines not found in the Culex homologue. Both Aedes genes were expressed in female salivary glands and in male mosquitoes (Arca et al. 2007, Ribeiro et al. 2007), but not in the carcass of female mosquitoes lacking the salivary glands, indicating that this family is specific of the salivary glands, but expressed also in males, suggesting a role in sugar feeding or as an antimicrobial peptide.
The 8.5-kDa Histidine-Rich Family
OT24, OT-27, and OT-28 are similar to proteins previously found in culicine sialotranscriptomes. These proteins have a histidine-rich domain in their mature amino termini, and >15 potential sites for O-galactosylations, suggesting they may function as mucins, and may also reduce the concentration of Zn because of their repeated histidine residues.
Proteins to Date Specific to Sialomes of Aedine Mosquitoes
The KKK Circle
A polypeptide deducted from the O. triseriatus sialotranscriptome matches (46% identity) a protein previously deducted from the Ae. albopictus sialotranscriptome (Arca et al. 2007) and annotated as glycine-rich peptide, but not found previously in other sialomes. It also matches a hypothetical protein from Ae. aegypti. Alignment of the three proteins (Fig. 7) shows higher sequence conservation at the carboxy-terminal end, where a triple lysine repeat is flanked by the only two cysteines found in the protein (Fig. 7, arrows), indicating the existence of an unusual structure of a highly positively charged ring. This protein family, unique to aedine mosquitoes to date, is hereby named the KKK circle. The Ae. albopictus transcript coding for this protein was found ubiquitously in mosquito tissues, suggesting a H role for this protein. Perhaps the basic charges drive this protein to be bound to negatively charged membrane phospholipids in the endoplasmic reticulum or Golgi (Murray et al. 1997).
Fig. 7.
The KKK circle family of aedine proteins. Clustal alignment of OT-492, an O. triseriatus protein, with Ae. aegypti and Ae. albopictus homologs. The bar under the alignment indicates a triple lysine surrounded by cysteines (arrows). (Online figure in color.)
The HHH Family
OT-16, OT-18, and OT-19 are homologues of previously identified proteins found in the sialotranscriptomes of Ae. aegypti and Ae. albopictus, which have a homopolymeric stretch of histidines near the mature amino termini. These proteins have no significant similarities to any other known protein. They may function as antimicrobials, as do many antimicrobial peptides that are histidine rich and may chelate Zn ions, a bacterial growth factor (Loomans et al. 1998).
Comparison of Protein Sequence Identities Between O. triseriatus and Ae. aegypti Gene Products
Ninety-three deduced protein sequences coding for putative H products are presented in Supplemental Table S2. These proteins allow comparison of the evolutionary rate of the S proteins compared with the H proteins, using the Ae. aegypti proteome (all Ae. aegypti proteins from GeneBank plus the proposed proteome found in VectorBase version AaegL1.1 from June 2006) as a reference set, as similarly done before for comparing An. stephensi salivary proteins to those of An. gambiae (Valenzuela et al. 2003). For this comparison, we used only protein sequences from O. triseriatus that had at least 80 amino acids of alignment by the blastp tool to a Ae. aegypti protein, and excluded from this set possible alleles or closely related gene duplications by removing the smaller sequence(s) that had 80% or more similarity to another one within the set, or any pair of proteins that matched the same Ae. aegypti protein, by removing the Ochlerotatus protein with the lowest identity. We thus compared 42 putative secreted O. triseriatus proteins with the Ae. aegypti proteome, obtaining an average of 64.2 + 2.5% protein identity (mean ± SEM), whereas 85 putative H proteins from O. triseriatus were 91.2 ± 2.5% identical to Ae. aegypti predicted proteins (P < 0.001 Mann-Whitney rank sum test). This significant difference further supports the concept that the evolution of mosquito salivary secreted proteins occurs at a faster pace than H proteins (Valenzuela et al. 2003, Calvo et al. 2007a, 2009b).
We have previously detected that the average codon volatility of salivary proteins was significantly larger than that of H genes (Calvo et al. 2010). It has been suggested that codon volatility (the proportion of the point-mutation neighbors of a codon that encode different amino acids) could be a measure of selection for fast evolution of proteins, as could occur in pathogens in a constant avoidance of antibody recognition (Friedman and Hughes 2005). Although this idea has created strong opposition (Chen et al. 2005, Pillai et al. 2005, Sharp 2005), it is supported by published models (Plotkin et al. 2006, Archetti 2009). We accordingly decided to measure the average codon volatility for the 85 sequences coding for H and 42 sequences coding for putative secreted proteins shown in Supplemental Table S2. The average codon volatility for the H class genes was 0.761869 ± 0.0011, whereas the S class had an average volatility of 0.7688 ± 0.0026 (average ± SEM), providing for a P value = 0.022 (double-tailed t test, nonhomogeneous variances). Whatever the discussion regarding the value of this index, it indicates that a single-point mutation on a S class gene has a significantly higher chance of producing a nonsynonymous amino acid substitution than in a H class gene.
Discussion
The sialosecretome of O. triseriatus conforms with those previously described by having transcripts coding for ubiquitous protein families, such as those coding for the enzymes amylase, maltase, chitinase, serine proteases, antiproteases of the serpin, Kazal and Trypsin inhibitor-like domain families, antigen-5 family, and immunity-related products associated with pathogen pattern recognition and antimicrobial functions. It also contains proteins unique to mosquitoes or other blood-sucking Nematocera, such as D7, 30-kDa/Aegyptin, 41-kDa, 56-kDa, basic tail, 13-kDa, 6.2-kDa, SG3/WW, and SGS families. The transcriptome analysis also identified members of the to date uniquely culicine families named 34-kDa, 30.5-kDa, WPCWW, and 8.5-kDa histidine-rich families, and the uniquely aedine HHH family.
The O. triseriatus sialotranscriptome also helped to identify partners to previously orphan mosquito sialome proteins, such as a secreted endonuclease, previously found only in the Cx. quinquefasciatus sialotranscriptome. Similarly, OT-157 helped to deorphanize a previously found protein from the Ae. aegypti sialome, with homologues found in the deducted proteomes of Cx. quinquefasciatus and An. gambiae. OT-547 deorphanized another Ae. aegypti sialo-protein, named the 8.5-kDa peptide, helping to identify previously taught to be an anopheline-specific family named the 6.2-kDa family (Fig. 4). OT-175 matched a W-rich sialoprotein from Ae. aegypti, from where a W-rich motif block was deducted and found to match previously described anopheline proteins of the SG3 family (Fig. 5). Finally, OT-492 matched an orphan sialoprotein from Ae. albopictus with a homologue found in the Ae. aegypti deducted proteome (but not in Anopheles or Culex proteomes), characterizing the aedine KKK circle family.
One protein family never found previously in sialotranscriptomes was identified in the O. triseriatus sialotranscriptome by nine ESTs coding for OT-148 and OT-150, constituting the GGHGQQ repeat family of glycine-rich peptides. Proteins of similar size and containing a signal peptide were also found in hypothetical proteins of Cx. quinquefasciatus and other insects, such as Nasonia vitripennis, but not in Aedes or Anopheles.
The O. triseriatus sialotranscriptome thus helped to consolidate the metasialome or sialoverse of mosquitoes in general, and that of culicines in particular, by adding to the pile of ubiquitously found mosquito proteins, by deorphanizing previously found odd proteins, by helping to create protein links between culicine and anopheline protein families, and by identification of a previously nondescribed protein family of insects never found before in mosquito sialomes. The comparison of the O. triseriatus sialome with that of Ae. aegypti also confirms the fast evolution of salivary proteins when compared with H ones, possibly because of host immune pressure.
Supplementary Material
Acknowledgments
This work was supported by the Intramural Research Program of the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health, and National Institutes of Health Grant R01 AI46435 to K.E.O.
References Cited
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ameri M, Wang X, Wilkerson MJ, Kanost MR, Broce AB. An immunoglobulin binding protein (antigen 5) of the stable fly (Diptera: Muscidae) salivary gland stimulates bovine immune responses. J Med Entomol. 2008;45:94–101. doi: 10.1603/0022-2585(2008)45[94:aibpao]2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arca B, Lombardo F, de Lara Capurro M, della Torre A, Dimopoulos G, James AA, Coluzzi M. Trapping cDNAs encoding secreted proteins from the salivary glands of the malaria vector Anopheles gambiae. Proc Natl Acad Sci USA. 1999;96:1516–1521. doi: 10.1073/pnas.96.4.1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arca B, Lombardo F, Valenzuela JG, Francischetti IM, Marinotti O, Coluzzi M, Ribeiro JM. An updated catalogue of salivary gland transcripts in the adult female mosquito, Anopheles gambiae. J Exp Biol. 2005;208:3971–3986. doi: 10.1242/jeb.01849. [DOI] [PubMed] [Google Scholar]
- Arca B, Lombardo F, Francischetti IM, Pham VM, Mestres-Simon M, Andersen JF, Ribeiro JM. An insight into the sialome of the adult female mosquito Aedes albopictus. Insect Biochem Mol Biol. 2007;37:107–127. doi: 10.1016/j.ibmb.2006.10.007. [DOI] [PubMed] [Google Scholar]
- Archetti M. Genetic robustness at the codon level as a measure of selection. Gene. 2009;443:64–69. doi: 10.1016/j.gene.2009.05.009. [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology: the Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman A, Birney E, Durbin R, Eddy SR, Howe KL, Sonnhammer EL. The Pfam protein families database. Nucleic Acids Res. 2000;28:263–266. doi: 10.1093/nar/28.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo E, Ribeiro JM. A novel secreted endonuclease from Culex quinquefasciatus salivary glands. J Exp Biol. 2006;209:2651–2659. doi: 10.1242/jeb.02267. [DOI] [PubMed] [Google Scholar]
- Calvo E, Mans BJ, Andersen JF, Ribeiro JM. Function and evolution of a mosquito salivary protein family. J Biol Chem. 2006;281:1935–1942. doi: 10.1074/jbc.M510359200. [DOI] [PubMed] [Google Scholar]
- Calvo E, Dao A, Pham VM, Ribeiro JM. An insight into the sialome of Anopheles funestus reveals an emerging pattern in anopheline salivary protein families. Insect Biochem Mol Biol. 2007a;37:164–175. doi: 10.1016/j.ibmb.2006.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo E, Tokumasu F, Marinotti O, Villeval JL, Ribeiro JM, Francischetti IM. Aegyptin, a novel mosquito salivary gland protein, specifically binds to collagen and prevents its interaction with platelet glycoprotein VI, integrin α2β1, and von Willebrand factor. J Biol Chem. 2007b;282:26928–26938. doi: 10.1074/jbc.M705669200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo E, Mans BJ, Ribeiro JM, Andersen JF. Multifunctionality and mechanism of ligand binding in a mosquito antiinflammatory protein. Proc Natl Acad Sci USA. 2009a;106:3728–3733. doi: 10.1073/pnas.0813190106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo E, Pham VM, Marinotti O, Andersen JF, Ribeiro JM. The salivary gland transcriptome of the neotropical malaria vector Anopheles darlingi reveals accelerated evolution of genes relevant to hematophagy. BMC Genomics. 2009b;10:57. doi: 10.1186/1471-2164-10-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo E, Sanchez-Vargas I, Favreau AJ, Barbian KD, Pham VM, Olson KE, Ribeiro JM. An insight into the sialotranscriptome of the West Nile mosquito vector, Culex tarsalis. BMC Genomics. 2010;11:51. doi: 10.1186/1471-2164-11-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champagne DE, Smartt CT, Ribeiro JM, James AA. The salivary gland-specific apyrase of the mosquito Aedes aegypti is a member of the 5′-nucleotidase family. Proc Natl Acad Sci USA. 1995;92:694–698. doi: 10.1073/pnas.92.3.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Emerson JJ, Martin TM. Evolutionary genomics: codon volatility does not detect selection. Nature. 2005;433:E6–E7. doi: 10.1038/nature03223. discussion E7–E8. [DOI] [PubMed] [Google Scholar]
- Craig GB., Jr Biology of Aedes triseriatus: some factors affecting control. Prog Clin Biol Res. 1983;123:329–341. [PubMed] [Google Scholar]
- DeFoliart GR. Aedes triseriatus: vector biology in relationship to the persistence of La Crosse virus in endemic foci. Prog Clin Biol Res. 1983;123:89–104. [PubMed] [Google Scholar]
- Edman J, Kale HW. Host behavior: its influence on the feeding success of mosquitoes. Ann Entomol Soc Am. 1971;64:513–516. [Google Scholar]
- Edman J, Webber LA, Schmid AA. Effect of host defenses on the feeding pattern of Culex nigripalpus when offered a choice of blood sources. J Parasitol. 1974;60:874–883. [PubMed] [Google Scholar]
- Edwards JF, Higgs S, Beaty BJ. Mosquito feeding-induced enhancement of Cache Valley virus (Bunyaviridae) infection in mice. J Med Entomol. 1998;35:261–265. doi: 10.1093/jmedent/35.3.261. [DOI] [PubMed] [Google Scholar]
- Friedman R, Hughes AL. Codon volatility as an indicator of positive selection: data from eukaryotic genome comparisons. Mol Biol Evol. 2005;22:542–546. doi: 10.1093/molbev/msi038. [DOI] [PubMed] [Google Scholar]
- Gibbs GM, Roelants K, O’Bryan MK. The CAP superfamily: cysteine-rich secretory proteins, antigen 5, and pathogenesis-related 1 proteins: roles in reproduction, cancer, and immune defense. Endocr Rev. 2008;29:865–897. doi: 10.1210/er.2008-0032. [DOI] [PubMed] [Google Scholar]
- Hekmat-Scafe DS, Dorit RL, Carlson JR. Molecular evolution of odorant-binding protein genes OS-E and OS-F in Drosophila. Genetics. 2000;155:117–127. doi: 10.1093/genetics/155.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Madan A. CAP3: a DNA sequence assembly program. Genome Res. 1999;9:868–877. doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isawa H, Yuda M, Orito Y, Chinzei Y. A mosquito salivary protein inhibits activation of the plasma contact system by binding to factor XII and high molecular weight kininogen. J Biol Chem. 2002;13:13. doi: 10.1074/jbc.M203505200. [DOI] [PubMed] [Google Scholar]
- Julenius K, Molgaard A, Gupta R, Brunak S. Prediction, conservation analysis, and structural characterization of mammalian mucin-type O-glycosylation sites. Glycobiology. 2005;15:153–164. doi: 10.1093/glycob/cwh151. [DOI] [PubMed] [Google Scholar]
- Korochkina S, Barreau C, Pradel G, Jeffery E, Li J, Natarajan R, Shabanowitz J, Hunt D, Frevert U, Vernick KD. A mosquito-specific protein family includes candidate receptors for malaria sporozoite invasion of salivary glands. Cell Microbiol. 2006;8:163–175. doi: 10.1111/j.1462-5822.2005.00611.x. [DOI] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Nei M. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- Loomans HJ, Hahn BL, Li QQ, Phadnis SH, Sohnle PG. Histidine-based zinc-binding sequences and the antimicrobial activity of calprotectin. J Infect Dis. 1998;177:812–814. doi: 10.1086/517816. [DOI] [PubMed] [Google Scholar]
- Ma D, Wang Y, Yang H, Wu J, An S, Gao L, Xu X, Lai R. Anti-thrombosis repertoire of blood-feeding horsefly salivary glands. Mol Cell Proteomics. 2009;8:2071–2079. doi: 10.1074/mcp.M900186-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mans BJ, Calvo E, Ribeiro JM, Andersen JF. The crystal structure of D7r4, a salivary biogenic amine-binding protein from the malaria mosquito Anopheles gambiae. J Biol Chem. 2007;282:36626–36633. doi: 10.1074/jbc.M706410200. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A, Panchenko AR, Shoemaker BA, Thiessen PA, Geer LY, Bryant SH. CDD: a database of conserved domain alignments with links to domain three-dimensional structure. Nucleic Acids Res. 2002;30:281–283. doi: 10.1093/nar/30.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellink JJ, Van Den Boven Kamp W. Functional aspects of mosquito salivation in blood feeding in Aedes aegypti. Mosq News. 1981;41:115–119. [Google Scholar]
- Milne TJ, Abbenante G, Tyndall JD, Halliday J, Lewis RJ. Isolation and characterization of a cone snail protease with homology to CRISP proteins of the pathogenesis-related protein superfamily. J Biol Chem. 2003;278:31105–31110. doi: 10.1074/jbc.M304843200. [DOI] [PubMed] [Google Scholar]
- Murray D, Ben-Tal N, Honig B, McLaughlin S. Electrostatic interaction of myristoylated proteins with membranes: simple physics, complicated biology. Structure. 1997;5:985–989. doi: 10.1016/s0969-2126(97)00251-7. [DOI] [PubMed] [Google Scholar]
- Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- Orlandi-Pradines E, Almeras L, Denis de Senneville L, Barbe S, Remoue F, Villard C, Cornelie S, Penhoat K, Pascual A, Bourgouin C, Fontenille D, Bonnet, Corre-Catelin N, Reiter P, Pages F, Laffite D, Boulanger D, Simondon F, Pradines B, Fusai T, Rogier C. Antibody response against saliva antigens of Anopheles gambiae and Aedes aegypti in travellers in tropical Africa. Microbes Infect. 2007;9:1454–1462. doi: 10.1016/j.micinf.2007.07.012. [DOI] [PubMed] [Google Scholar]
- Peng Z, Li H, Simons FE. Immunoblot analysis of salivary allergens in 10 mosquito species with worldwide distribution and the human IgE responses to these allergens. J Allergy Clin Immunol. 1998;101:498–505. doi: 10.1016/S0091-6749(98)70357-4. [DOI] [PubMed] [Google Scholar]
- Pillai SK, Kosakovsky Pond SL, Woelk CH, Richman DD, Smith DM. Codon volatility does not reflect selective pressure on the HIV-1 genome. Virology. 2005;336:137–143. doi: 10.1016/j.virol.2005.03.014. [DOI] [PubMed] [Google Scholar]
- Plotkin JB, Dushoff J, Desai MM, Fraser HB. Codon usage and selection on proteins. J Mol Evol. 2006;63:635–653. doi: 10.1007/s00239-005-0233-x. [DOI] [PubMed] [Google Scholar]
- Reinert JF. New classification for the composite genus Aedes (Diptera: Culicidae: Aedini), elevation of subgenus Ochlerotatus to generic rank, reclassification of the other subgenera, and notes on certain subgenera and species. J Am Mosq Control Assoc. 2000;16:175–188. [PubMed] [Google Scholar]
- Reno HE, Novak RJ. Characterization of apyrase-like activity in Ochlerotatus triseriatus, Ochlerotatus hendersoni, and Aedes aegypti. Am J Trop Med Hyg. 2005;73:541–545. [PubMed] [Google Scholar]
- Ribeiro JMC, Arca B. From sialomes to the sialoverse: an insight into the salivary potion of blood feeding insects. Adv Insect Physiol. 2009;37:59–118. [Google Scholar]
- Ribeiro JM, Francischetti IM. Role of arthropod saliva in blood feeding: sialome and post-sialome perspectives. Annu Rev Entomol. 2003;48:73–88. doi: 10.1146/annurev.ento.48.060402.102812. [DOI] [PubMed] [Google Scholar]
- Ribeiro JM, Nussenzveig RH, Tortorella G. Salivary vasodilators of Aedes triseriatus and Anopheles gambiae (Diptera: Culicidae) J Med Entomol. 1994;31:747–753. doi: 10.1093/jmedent/31.5.747. [DOI] [PubMed] [Google Scholar]
- Ribeiro JM, Charlab R, Pham VM, Garfield M, Valenzuela JG. An insight into the salivary transcriptome and proteome of the adult female mosquito Culex pipiens quinquefasciatus. Insect Biochem Mol Biol. 2004;34:543–563. doi: 10.1016/j.ibmb.2004.02.008. [DOI] [PubMed] [Google Scholar]
- Ribeiro JM, Arca B, Lombardo F, Calvo E, Phan VM, Chandra PK, Wikel SK. An annotated catalogue of salivary gland transcripts in the adult female mosquito, Aedes aegypti. BMC Genomics. 2007;8:6. doi: 10.1186/1471-2164-8-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero JR, Newland JG. Viral meningitis and encephalitis: traditional and emerging viral agents. Semin Pediatr Infect Dis. 2003;14:72–82. doi: 10.1053/spid.2003.127223. [DOI] [PubMed] [Google Scholar]
- Rossignol PA, Ribeiro JM, Spielman A. Increased intradermal probing time in sporozoite-infected mosquitoes. Am J Trop Med Hyg. 1984;33:17–20. doi: 10.4269/ajtmh.1984.33.17. [DOI] [PubMed] [Google Scholar]
- Rust RS, Thompson WH, Matthews CG, Beaty BJ, Chun RW. La Crosse and other forms of California encephalitis. J Child Neurol. 1999;14:1–14. doi: 10.1177/088307389901400101. [DOI] [PubMed] [Google Scholar]
- Schultz J, Copley RR, Doerks T, Ponting CP, Bork P. SMART: a web-based tool for the study of genetically mobile domains. Nucleic Acids Res. 2000;28:231–234. doi: 10.1093/nar/28.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp PM. Gene “volatility” is most unlikely to reveal adaptation. Mol Biol Evol. 2005;22:807–809. doi: 10.1093/molbev/msi073. [DOI] [PubMed] [Google Scholar]
- Shi L, Paskewitz SM. Identification and molecular characterization of two immune-responsive chitinase-like proteins from Anopheles gambiae. Insect Mol Biol. 2004;13:387–398. doi: 10.1111/j.0962-1075.2004.00496.x. [DOI] [PubMed] [Google Scholar]
- Stark KR, James AA. Isolation and characterization of the gene encoding a novel factor Xa-directed anticoagulant from the yellow fever mosquito, Aedes aegypti. J Biol Chem. 1998;273:20802–20809. doi: 10.1074/jbc.273.33.20802. [DOI] [PubMed] [Google Scholar]
- Sun D, McNicol A, James AA, Peng Z. Expression of functional recombinant mosquito salivary apyrase: a potential therapeutic platelet aggregation inhibitor. Platelets. 2006;17:178–184. doi: 10.1080/09537100500460234. [DOI] [PubMed] [Google Scholar]
- Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov SL, Nikolskaya AN, Rao BS, Smirnov S, Sverdlov AV, Vasudevan S, Wolf YI, Yin JJ, Natale DA. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzuela JG, Pham VM, Garfield MK, Francischetti IM, Ribeiro JMC. Toward a description of the sialome of the adult female mosquito Aedes aegypti. Insect Biochem Mol Biol. 2002a;32:1101–1122. doi: 10.1016/s0965-1748(02)00047-4. [DOI] [PubMed] [Google Scholar]
- Valenzuela JG, Charlab R, Gonzalez EC, Miranda-Santos IKF, Marinotti O, Francischetti IM, Ribeiro JMC. The D7 family of salivary proteins in blood sucking Diptera. Insect Mol Biol. 2002b;11:149–155. doi: 10.1046/j.1365-2583.2002.00319.x. [DOI] [PubMed] [Google Scholar]
- Valenzuela JG, Francischetti IM, Pham VM, Garfield MK, Ribeiro JM. Exploring the salivary gland transcriptome and proteome of the Anopheles stephensi mosquito. Insect Biochem Mol Biol. 2003;33:717–732. doi: 10.1016/s0965-1748(03)00067-5. [DOI] [PubMed] [Google Scholar]
- Vizioli J, Bulet P, Hoffmann JA, Kafatos FC, Muller HM, Dimopoulos G. Gambicin: a novel immune responsive antimicrobial peptide from the malaria vector Anopheles gambiae. Proc Natl Acad Sci USA. 2001;98:12630–12635. doi: 10.1073/pnas.221466798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler DL, Barrett T, Benson DA, Bryant SH, Canese K, Church DM, DiCuccio M, Edgar R, Federhen S, Helmberg W, Kenton DL, Khovayko O, Lipman DJ, Madden TL, Maglott DR, Ostell J, Pontius JU, Pruitt KD, Schuler GD, Schriml LM, Sequeira E, Sherry ST, Sirotkin K, Starchenko G, Suzek TO, Tatusov R, Tatusova TA, Wagner L, Yaschenko E. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2005;33:D39–D45. doi: 10.1093/nar/gki062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Yang H, Ma D, Wu J, Wang Y, Song Y, Wang X, Lu Y, Yang J, Lai R. Toward an understanding of the molecular mechanism for successful blood feeding by coupling proteomics analysis with pharmacological testing of horsefly salivary glands. Mol Cell Proteomics. 2008;7:582–590. doi: 10.1074/mcp.M700497-MCP200. [DOI] [PubMed] [Google Scholar]
- Yamazaki Y, Hyodo F, Morita T. Wide distribution of cysteine-rich secretory proteins in snake venoms: isolation and cloning of novel snake venom cysteine-rich secretory proteins. Arch Biochem Biophys. 2003;412:133–141. doi: 10.1016/s0003-9861(03)00028-6. [DOI] [PubMed] [Google Scholar]
- Yoshida S, Sudo T, Niimi M, Tao L, Sun B, Kambayashi J, Watanabe H, Luo E, Matsuoka H. Inhibition of collagen-induced platelet aggregation by anopheline antiplatelet protein, a saliva protein from a malaria vector mosquito. Blood. 2008;111:2007–2014. doi: 10.1182/blood-2007-06-097824. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.